Childhood Cancer Genomics (PDQ®): Treatment - Health Professional Information [NCI]
Childhood Cancer Genomics (PDQ®): Treatment - Health Professional Information [NCI]Skip to the navigationGeneral Information About Childhood Cancer GenomicsResearch teams from around the world have made remarkable progress in the past decade in elucidating the genomic landscape of most types of childhood cancer. A decade ago it was possible to hope that targetable oncogenes, such as activated tyrosine kinases, might be identified in a high percentage of childhood cancers. However, it is now clear that the genomic landscape of childhood cancer is highly varied, and in many cases is quite distinctive from that of the common adult cancers. There are examples of genomic lesions that have provided immediate therapeutic direction, including the following: - NPM-ALK fusion genes associated with anaplastic large cell lymphoma cases.
- ALK point mutations associated with a subset of neuroblastoma cases.
- BRAF and other kinase genomic alterations associated with subsets of pediatric glioma cases.
- Hedgehog pathway mutations associated with a subset of medulloblastoma cases.
- ABL family genes activated by translocation in a subset of acute lymphoblastic leukemia (ALL) cases.
For some cancers, the genomic findings have been highly illuminating in the identification of genomically defined subsets of patients within histologies that have distinctive biological features and distinctive clinical characteristics (particularly in terms of prognosis). In some instances, identification of these subtypes has resulted in early clinical translation as exemplified by the WNT subgroup of medulloblastoma. Because of its excellent outcome, the WNT subgroup will be studied separately in future medulloblastoma clinical trials so that reductions in therapy can be evaluated with the goal of maintaining favorable outcome while reducing long-term morbidity. However, the prognostic significance of the recurring genomic lesions for some other cancers remains to be defined. A key finding from genomic studies is the extent to which the molecular characteristics of childhood cancers correlate with their tissue (cell) of origin. As with most adult cancers, mutations in childhood cancers do not arise at random, but rather are linked in specific constellations to disease categories. A few examples include the following: - The presence of H3.3 and H3.1 K27 mutations almost exclusively among pediatric midline high-grade gliomas.
- The loss of SMARCB1 in rhabdoid tumors.
- The presence of RELA translocations in supratentorial ependymomas.
- The presence of specific fusion proteins in different pediatric sarcomas.
Another theme across multiple childhood cancers is the contribution of mutations of genes involved in normal development of the tissue of origin of the cancer and the contribution of genes involved in epigenomic regulation. Structural variations play an important role for many childhood cancers. Translocations resulting in oncogenic fusion genes or overexpression of oncogenes play a central role, particularly for the leukemias and sarcomas. However, for other childhood cancers that are primarily characterized by structural variations, functional fusion genes are not produced. Mechanisms by which these recurring structural variations have oncogenic effects have been identified for osteosarcoma (translocations confined to the first intron of TP53) and medulloblastoma (structural variants juxtapose GFI1 or GFI1B coding sequences proximal to active enhancer elements leading to transcriptional activation [enhancer hijacking]).[1,2] However, the oncogenic mechanisms of action for recurring structural variations of other childhood cancers (e.g., the segmental chromosomal alterations in neuroblastoma) need to be elucidated. Understanding of the contribution of germline mutations to childhood cancer etiology is being advanced by the application of whole-genome and exome sequencing to cohorts of children with cancer. Estimates for rates of pathogenic germline mutations approaching 10% have emerged from studies applying these sequencing methods to childhood cancer cohorts.[3,4,5] In some cases, the pathogenic germline mutations are clearly contributory to the patient's cancer (e.g., TP53 mutations arising in the context of Li-Fraumeni syndrome), whereas in other cases the contribution of the germline mutation to the patient's cancer is less clear (e.g., mutations in adult cancer predisposition genes such as BRCA1 and BRCA2 that have an undefined role in childhood cancer predisposition).[4,5] The frequency of germline mutations varies by tumor type (e.g., lower for neuroblastoma and higher for osteosarcoma),[5] and many of the identified germline mutations fit into known predisposition syndromes (e.g., DICER1 for pleuropulmonary blastoma, SMARCB1 and SMARCA4 for rhabdoid tumor and small cell ovarian cancer, TP53 for adrenocortical carcinoma and Li-Fraumeni syndrome cancers, RB1 for retinoblastoma, etc.). The germline contribution to the development of specific cancers is discussed in the disease-specific sections that follow. Each section of this document is meant to provide readers with a brief summary of current knowledge about the genomic landscape of specific childhood cancers, an understanding that is critical in considering how to apply precision medicine concepts to childhood cancers. References:
-
Northcott PA, Lee C, Zichner T, et al.: Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511 (7510): 428-34, 2014.
-
Chen X, Bahrami A, Pappo A, et al.: Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep 7 (1): 104-12, 2014.
-
Mody RJ, Wu YM, Lonigro RJ, et al.: Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA 314 (9): 913-25, 2015.
-
Parsons DW, Roy A, Yang Y, et al.: Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol : , 2016.
-
Zhang J, Walsh MF, Wu G, et al.: Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med 373 (24): 2336-46, 2015.
LeukemiasAcute Lymphoblastic Leukemia (ALL) The genomics of childhood ALL has been extensively investigated and multiple distinctive subtypes based on cytogenetic and molecular characterizations have been defined, each with its own pattern of clinical and prognostic characteristics.[1] Figure 1 illustrates the distribution of ALL cases by cytogenetic/molecular subtype.[1]
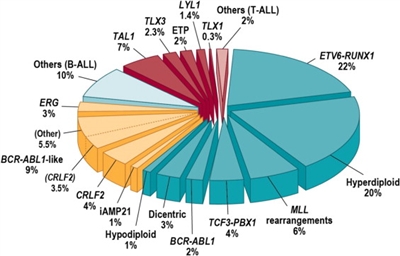
Figure 1. Subclassification of childhood ALL. Blue wedges refer to B-progenitor ALL, yellow to recently identified subtypes of B-ALL, and red wedges to T-lineage ALL. Reprinted from Seminars in Hematology, Volume 50, Charles G. Mullighan, Genomic Characterization of Childhood Acute Lymphoblastic Leukemia, Pages 314-324, Copyright (2013), with permission from Elsevier. The genomic landscape of B-precursor ALL is typified by a range of genomic alterations that disrupt normal B-cell development and in some cases by mutations in genes that provide a proliferation signal (e.g., activating mutations in RAS family genes or mutations/translocations leading to kinase pathway signaling). Genomic alterations leading to blockage of B-cell development include translocations (e.g., TCF3-PBX1 and ETV6-RUNX1), point mutations (e.g., IKZF1 and PAX5), and intragenic/intergenic deletions (e.g., IKZF1, PAX5, EBF, and ERG).[2] The genomic alterations in B-precursor ALL tend not to occur at random, but rather to cluster within subtypes that can be delineated by biological characteristics such as their gene expression profiles. Cases with recurring chromosomal translocations (e.g., TCF3-PBX1 and ETV6-RUNX1, and MLL (KMT2A)-rearranged ALL) have distinctive biological features and illustrate this point, as do the examples below of specific genomic alterations within distinctive biological subtypes: - IKZF1 deletions and mutations are most commonly observed within cases of Philadelphia chromosome-positive (Ph+) ALL and Ph-like ALL.[3,4]
- Intragenic ERG deletions occur within a distinctive subtype characterized by this alteration and lacking other recurring cytogenetic alterations associated with pediatric B-precursor ALL.[5,6,7]
- TP53 mutations occur at high frequency in patients with low hypodiploid ALL with 32 to 39 chromosomes, and the TP53 mutations in these patients are often germline.[8]TP53 mutations are uncommon in other patients with B-precursor ALL.
Activating point mutations in kinase genes are uncommon in high-risk B-precursor ALL, and JAK genes are the primary kinases that are found to be mutated. These mutations are generally observed in patients with Ph-like ALL that have CRLF2 abnormalities, although JAK2 mutations are also observed in approximately 15% of children with Down syndrome ALL.[4,9,10] Several kinase genes and cytokine receptor genes are activated by translocation as described below in the discussion of Ph-positive ALL and Ph-like ALL. FLT3 mutations occur in a minority of cases (approximately 10%) of hyperdiploid ALL and MLL (KMT2A)-rearranged ALL, and are rare in other subtypes.[11] Understanding of the genomics of B-precursor ALL at relapse is less advanced than understanding of ALL genomics at diagnosis. Childhood ALL is often polyclonal at diagnosis and under the selective influence of therapy, some clones may be extinguished and new clones with distinctive genomic profiles may arise.[12] Of particular importance are new mutations that arise at relapse that may be selected by specific components of therapy. As an example, mutations in NT5C2 are not found at diagnosis whereas specific mutations in NT5C2 were observed in 7 of 44 (16%) and 9 of 20 (45%) cases of B-precursor ALL with early relapse that were evaluated for this mutation.[12,13]NT5C2 mutations are uncommon in patients with late relapse, and they appear to induce resistance to 6-mercaptopurine and thioguanine.[13] Another gene that is found mutated only at relapse is PRSP1, a gene involved purine biosynthesis.[14] Mutations were observed in 13.0% of a Chinese cohort and 2.7% of a German cohort, and were observed in patients with on-treatment relapses. The PRSP1 mutations observed in relapsed cases induce resistance to thiopurines in leukemia cell lines. CREBBP mutations are also enriched at relapse and appear to be associated with increased resistance to glucocorticoids.[12,15] With increased understanding of the genomics of relapse, it may be possible to tailor upfront therapy to avoid relapse or detect resistance-inducing mutations early and intervene before a frank relapse. Specific genomic and chromosomal alterations are provided below, with a focus on their prognostic significance. A number of recurrent chromosomal abnormalities have been shown to have prognostic significance, especially in precursor B-cell ALL. Some chromosomal alterations are associated with more favorable outcomes, such as high hyperdiploidy (51-65 chromosomes) and the ETV6-RUNX1 fusion. Others historically have been associated with a poorer prognosis, including the Philadelphia chromosome (t(9;22)(q34;q11.2)), rearrangements of the MLL (KMT2A) gene, hypodiploidy, and intrachromosomal amplification of the AML1 gene (iAMP21).[16] In recognition of the clinical significance of many of these genomic alterations, the 2016 revision of the World Health Organization classification of tumors of the hematopoietic and lymphoid tissues lists the following entities for precursor B-cell ALL:[17] - B-lymphoblastic leukemia/lymphoma, not otherwise specified (NOS).
- B-lymphoblastic leukemia/lymphoma with recurrent genetic abnormalities.
- B-lymphoblastic leukemia/lymphoma with t(9;22)(q34.1;q11.2); BCR-ABL1.
- B-lymphoblastic leukemia/lymphoma with t(v;11q23.3); KMT2A rearranged.
- B-lymphoblastic leukemia/lymphoma with t(12;21)(p13.2;q22.1); ETV6-RUNX1.
- B-lymphoblastic leukemia/lymphoma with hyperdiploidy.
- B-lymphoblastic leukemia/lymphoma with hypodiploidy.
- B-lymphoblastic leukemia/lymphoma with t(5;14)(q31.1;q32.3); IL3-IGH.
- B-lymphoblastic leukemia/lymphoma with t(1;19)(q23;p13.3); TCF3-PBX1.
- Provisional entity: B-lymphoblastic leukemia/lymphoma, BCR-ABL1-like.
- Provisional entity: B-lymphoblastic leukemia/lymphoma with iAMP21.
These and other chromosomal and genomic abnormalities for childhood ALL are described below. - Chromosome number
- High hyperdiploidy (51-65 chromosomes)
High hyperdiploidy, defined as 51 to 65 chromosomes per cell or a DNA index greater than 1.16, occurs in 20% to 25% of cases of precursor B-cell ALL, but very rarely in cases of T-cell ALL.[18] Hyperdiploidy can be evaluated by measuring the DNA content of cells (DNA index) or by karyotyping. In cases with a normal karyotype or in which standard cytogenetic analysis was unsuccessful, interphase fluorescence in situ hybridization (FISH) may detect hidden hyperdiploidy. High hyperdiploidy generally occurs in cases with clinically favorable prognostic factors (patients aged 1 to <10 years with a low white blood cell [WBC] count) and is itself an independent favorable prognostic factor.[18,19,20] Within the hyperdiploid range of 51 to 65 chromosomes, patients with higher modal numbers (58-66) appeared to have a better prognosis in one study.[20] Hyperdiploid leukemia cells are particularly susceptible to undergoing apoptosis and accumulate higher levels of methotrexate and its active polyglutamate metabolites,[21] which may explain the favorable outcome commonly observed in these cases. While the overall outcome of patients with high hyperdiploidy is considered to be favorable, factors such as age, WBC count, specific trisomies, and early response to treatment have been shown to modify its prognostic significance.[22,23] Patients with trisomies of chromosomes 4, 10, and 17 (triple trisomies) have been shown to have a particularly favorable outcome as demonstrated by both Pediatric Oncology Group (POG) and Children's Cancer Group analyses of NCI standard-risk ALL.[24] POG data suggest that NCI standard-risk patients with trisomies of 4 and 10, without regard to chromosome 17 status, have an excellent prognosis.[25] Chromosomal translocations may be seen with high hyperdiploidy, and in those cases, patients are more appropriately risk-classified based on the prognostic significance of the translocation. For instance, in one study, 8% of patients with the Philadelphia chromosome (t(9;22)(q34;q11.2)) also had high hyperdiploidy,[26] and the outcome of these patients (treated without tyrosine kinase inhibitors) was inferior to that observed in non-Philadelphia chromosome-positive (Ph+) high hyperdiploid patients. Certain patients with hyperdiploid ALL may have a hypodiploid clone that has doubled (masked hypodiploidy).[27] These cases may be interpretable based on the pattern of gains and losses of specific chromosomes. These patients have an unfavorable outcome, similar to those with hypodiploidy.[27] Near triploidy (68-80 chromosomes) and near tetraploidy (>80 chromosomes) are much less common and appear to be biologically distinct from high hyperdiploidy.[28] Unlike high hyperdiploidy, a high proportion of near tetraploid cases harbor a cryptic ETV6-RUNX1 fusion.[28,29,30] Near triploidy and tetraploidy were previously thought to be associated with an unfavorable prognosis, but later studies suggest that this may not be the case.[28,30] The genomic landscape of hyperdiploid ALL is represented by mutations in genes of the receptor tyrosine kinase (RTK)/RAS pathway in approximately one-half of cases. Genes encoding histone modifiers are also present in a recurring manner in a minority of cases. Analysis of mutation profiles demonstrates that chromosomal gains are early events in the pathogenesis of hyperdiploid ALL.[31] - Hypodiploidy (<44 chromosomes)
Precursor B-cell ALL cases with fewer than the normal number of chromosomes have been subdivided in various ways, with one report stratifying based on modal chromosome number into the following four groups:[27] - Near-haploid: 24 to 29 chromosomes (n = 46).
- Low-hypodiploid: 33 to 39 chromosomes (n = 26).
- High-hypodiploid: 40 to 43 chromosomes (n = 13).
- Near-diploid: 44 chromosomes (n = 54).
Most patients with hypodiploidy are in the near-haploid and low-hypodiploid groups, and both of these groups have an elevated risk of treatment failure compared with nonhypodiploid cases.[27,32] Patients with fewer than 44 chromosomes have a worse outcome than do patients with 44 or 45 chromosomes in their leukemic cells.[27] One study of 20 patients with near-haploid or low-hypodiploid ALL indicated that MRD may have prognostic significance in the hypodiploid population.[33] The recurring genomic alterations of near-haploid and low-hypodiploid ALL appear to be distinctive from each other and from other types of ALL.[8] In near-haploid ALL, alterations targeting RTK signaling, RAS signaling, and IKZF3 are common.[34] In low-hypodiploid ALL, genetic alterations involving TP53, RB1, and IKZF2 are common. Importantly, the TP53 alterations observed in low-hypodiploid ALL are also present in nontumor cells in approximately 40% of cases, suggesting that these mutations are germline and that low-hypodiploid ALL represents, in some cases, a manifestation of Li-Fraumeni syndrome.[8]
- Chromosomal translocations and gains/deletions of chromosomal segments
- t(12;21)(p13.2;q22.1); ETV6-RUNX1 (formerly known as TEL-AML1)
Fusion of the ETV6 gene on chromosome 12 to the RUNX1 gene on chromosome 21 is present in 20% to 25% of cases of precursor B-cell ALL but is rarely observed in T-cell ALL.[29] The t(12;21)(p12;q22) produces a cryptic translocation that is detected by methods such as FISH, rather than conventional cytogenetics, and it occurs most commonly in children aged 2 to 9 years.[35,36] Hispanic children with ALL have a lower incidence of t(12;21)(p13;q22) than do white children.[37] Reports generally indicate favorable event-free survival (EFS) and overall survival (OS) in children with the ETV6-RUNX1 fusion; however, the prognostic impact of this genetic feature is modified by the following factors:[38,39,40,41,42] - Early response to treatment.
- NCI risk category (age and WBC count at diagnosis).
- Treatment regimen.
In one study of the treatment of newly diagnosed children with ALL, multivariate analysis of prognostic factors found age and leukocyte count, but not ETV6-RUNX1, to be independent prognostic factors.[38] It does not appear that the presence of secondary cytogenetic abnormalities, such as deletion of ETV6 (12p) or CDKN2A/B (9p), impacts the outcome of patients with the ETV6-RUNX1 fusion.[42,43] There is a higher frequency of late relapses in patients with ETV6-RUNX1 fusion compared with other precursor B-cell ALL.[38,44] Patients with the ETV6-RUNX1 fusion who relapse seem to have a better outcome than other relapse patients,[45] with an especially favorable prognosis for patients who relapse more than 36 months from diagnosis.[46] Some relapses in patients with t(12;21)(p13;q22) may represent a new independent second hit in a persistent preleukemic clone (with the first hit being the ETV6-RUNX1 translocation).[47,48] - t(9;22)(q34.1;q11.2); BCR-ABL1 (Ph+)
The Philadelphia chromosome t(9;22)(q34;q11.2) is present in approximately 3% of children with ALL and leads to production of a BCR-ABL1 fusion protein with tyrosine kinase activity (refer to Figure 2).
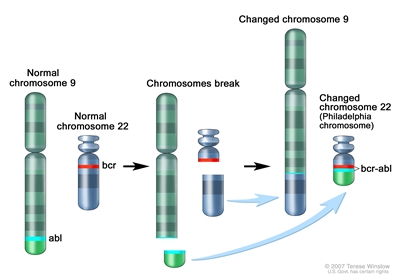
Figure 2. The Philadelphia chromosome is a translocation between the ABL-1 oncogene (on the long arm of chromosome 9) and the breakpoint cluster region (BCR) (on the long arm of chromosome 22), resulting in the fusion gene BCR-ABL1. BCR-ABL1 encodes an oncogenic protein with tyrosine kinase activity. This subtype of ALL is more common in older children with precursor B-cell ALL and high WBC count, with the incidence of the t(9;22)(q34;q11.2) increasing to about 25% in young adults with ALL. Historically, the Philadelphia chromosome t(9;22)(q34;q11.2) was associated with an extremely poor prognosis (especially in those who presented with a high WBC count or had a slow early response to initial therapy), and its presence had been considered an indication for allogeneic hematopoietic stem cell transplantation (HSCT) in patients in first remission.[26,49,50,51] Inhibitors of the BCR-ABL1 tyrosine kinase, such as imatinib mesylate, are effective in patients with Ph+ ALL.[52] A study by the Children's Oncology Group (COG), which used intensive chemotherapy and concurrent imatinib mesylate given daily, demonstrated a 5-year EFS rate of 70% ± 12%, which was superior to the EFS rate of historical controls in the pre-tyrosine kinase inhibitor (imatinib mesylate) era.[53,54] - t(v;11q23.3); MLL (KMT2A)-rearranged
Rearrangements involving the MLL (KMT2A) gene occur in approximately 5% of childhood ALL cases overall, but in up to 80% of infants with ALL. These rearrangements are generally associated with an increased risk of treatment failure.[55,56,57,58] The t(4;11)(q21;q23) is the most common rearrangement involving the MLL gene in children with ALL and occurs in approximately 1% to 2% of childhood ALL.[56,59] Patients with the t(4;11)(q21;q23) are usually infants with high WBC counts; they are more likely than other children with ALL to have central nervous system (CNS) disease and to have a poor response to initial therapy.[60] While both infants and adults with the t(4;11)(q21;q23) are at high risk of treatment failure, children with the t(4;11)(q21;q23) appear to have a better outcome than either infants or adults.[55,56] Irrespective of the type of MLL (KMT2A) gene rearrangement, infants with leukemia cells that have MLL gene rearrangements have a worse treatment outcome than older patients whose leukemia cells have an MLL gene rearrangement.[55,56] Whole-genome sequencing has determined that cases of infant ALL with MLL gene rearrangements have few additional genomic alterations, none of which have clear clinical significance.[11] Deletion of the MLL gene has not been associated with an adverse prognosis.[61] Of interest, the t(11;19)(q23;p13.3) involving MLL (KMT2A) and MLLT1/ENL occurs in approximately 1% of ALL cases and occurs in both early B-lineage and T-cell ALL.[62] Outcome for infants with the t(11;19) is poor, but outcome appears relatively favorable in older children with T-cell ALL and the t(11;19).[62] - t(1;19)(q23;p13.3); TCF3-PBX1 and t(17;19)(q22;p13); TCF3-HLF
The t(1;19) occurs in approximately 5% of childhood ALL cases and involves fusion of the TCF3 gene on chromosome 19 to the PBX1 gene on chromosome 1.[63,64] The t(1;19) may occur as either a balanced translocation or as an unbalanced translocation and is the primary recurring genomic alteration of the pre-B ALL immunophenotype (cytoplasmic Ig positive).[65] Black children are relatively more likely than white children to have pre-B ALL with the t(1;19).[66] The t(1;19) had been associated with inferior outcome in the context of antimetabolite-based therapy,[67] but the adverse prognostic significance was largely negated by more aggressive multiagent therapies.[64,68] However, in a trial conducted by St. Jude Children's Research Hospital (SJCRH) on which all patients were treated without cranial radiation, patients with the t(1;19) had an overall outcome comparable to children lacking this translocation, with a higher risk of CNS relapse and a lower rate of bone marrow relapse, suggesting that more intensive CNS therapy may be needed for these patients.[69,70] The t(17;19) resulting in the TCF3-HLF fusion occurs in less than 1% of pediatric ALL cases. ALL with the TCF3-HLF fusion is associated at diagnosis with disseminated intravascular coagulation and with hypercalcemia. Outcome is very poor for children with the t(17;19), with a literature review noting mortality for 20 of 21 cases reported.[71] In addition to the TCF3-HLF fusion, the genomic landscape of this ALL subtype was characterized by deletions in genes involved in B-cell development (PAX5, BTG1, and VPREB1) and by mutations in RAS pathway genes (NRAS, KRAS, and PTPN11).[65] - DUX4-rearranged ALL with frequent ERG deletions
Approximately 5% of standard-risk and 10% of high-risk pediatric precursor B-cell ALL patients have a rearrangement involving DUX4 that leads to its overexpression.[72,73] The frequency in older adolescents (aged >15 years) is approximately 10%. The most common rearrangement produces IGH-DUX4 fusions, with ERG-DUX4 fusions also observed. Approximately 50% of DUX4-rearranged cases have focal intragenic deletions involving ERG that are not observed in other ALL subtypes,[72,73] and DUX4-rearranged cases show a distinctive gene expression pattern that was initially identified as being associated with these focal deletions in ERG.[5,6,7]IKZF1 alterations are observed in 35% to 40% of DUX4-rearranged ALL.[72,73]ERG deletion connotes an excellent prognosis, with OS exceeding 90%; even when the IZKF1 deletion is present, prognosis remains highly favorable.[5,6,7] Patients with DUX4 rearrangements who lack ERG deletion also appear to have favorable prognosis.[73] - t(5;14)(q31.1;q32.3); IL3-IGH
This entity is included in the 2016 revision of the WHO classification of tumors of the hematopoietic and lymphoid tissues.[17] The finding of t(5;14)(q31.1;q32.3) in patients with ALL and hypereosinophilia in the 1980s was followed by the identification of the IL3-IGH fusion as the underlying genetic basis for the condition.[74,75] The joining of the IGH locus to the promoter region of the interleukin-3 gene (IL3) leads to dysregulation of IL3 expression.[76] Cytogenetic abnormalities in children with ALL and eosinophilia are variable, with only a subset resulting from the IL3-IGH fusion.[77] The number of cases of IL3-IGH ALL described in the published literature is too small to assess the prognostic significance of the IL3-IGH fusion. - Intrachromosomal amplification of chromosome 21 (iAMP21)
iAMP21 with multiple extra copies of the RUNX1 (AML1) gene at 21q22 occurs in approximately 2% of precursor B-cell ALL cases and is associated with older age (median, approximately 10 years), presenting WBC of less than 50 × 109 /L, a slight female preponderance, and high end-induction minimal residual disease (MRD).[78,79,80] The United Kingdom (UK)-ALL clinical trials group initially reported that the presence of iAMP21 conferred a poor prognosis in patients treated in the MRC ALL 97/99 trial (5-year EFS, 29%).[16] In their subsequent trial (UKALL2003 [NCT00222612]), patients with iAMP21 were assigned to a more intensive chemotherapy regimen and had a markedly better outcome (5-year EFS, 78%).[79] Similarly, the COG has reported that iAMP21 was associated with a significantly inferior outcome in NCI standard-risk patients (4-year EFS, 73% for iAMP21 vs. 92% in others), but not in NCI high-risk patients (4-year EFS, 73% vs. 80%).[78] On multivariate analysis, iAMP21 was an independent predictor of inferior outcome only in NCI standard-risk patients.[78] The results of the UKALL2003 and COG studies suggest that treatment of iAMP21 patients with high-risk chemotherapy regimens abrogates its adverse prognostic significance and obviates the need for SCT in first remission.[80] - IKZF1 deletions
IKZF1 deletions, including deletions of the entire gene and deletions of specific exons, are present in approximately 15% of precursor B-cell ALL cases. Less commonly, IKZF1 can be inactivated by deleterious point mutations.[81] Cases with IKZF1 deletions tend to occur in older children, have a higher WBC count at diagnosis, and are therefore, more common in NCI high-risk patients than in NCI standard-risk patients.[2,81,82,83] A high proportion of BCR-ABL1 cases have a deletion of IKZF1,[3,82] and ALL arising in children with Down syndrome appears to have elevated rates of IKZF1 deletions.[84]IKZF1 deletions are also common in cases with CRLF2 genomic alterations and in Philadelphia chromosome (Ph)-like (BCR-ABL1-like) ALL (see below).[5,82,85] Multiple reports have documented the adverse prognostic significance of an IKZF1 deletion, and most studies have reported that this deletion is an independent predictor of poor outcome on multivariate analyses.[5,81,82,85,86,87,88,89,90,91]; [92][Level of evidence: 2Di] That said, the prognostic significance of IKZF1 may not apply equally across ALL biological subtypes, as illustrated by the apparent lack of prognostic significance in patients with ERG deletion.[7] - BCR-ABL1-like (Ph-like)
BCR-ABL1-negative patients with a gene expression profile similar to BCR-ABL1-positive patients have been referred to as BCR-ABL1-like.[81,85] This occurs in 10% to 15% of pediatric ALL patients, increasing in frequency with age, and has been associated with a poor prognosis and with IKZF1 deletion or mutation.[9,81,85,89,93] The 5-year EFS rate of 90% observed in one study of 40 patients with BCR-ABL1-like ALL suggested that the adverse prognostic significance of this subtype may be abrogated when patients are treated with risk-directed therapy based on MRD levels. Six of these 40 patients were classified as high risk and all proceeded to allogeneic SCT.[94][Level of evidence: 2A] The hallmark of BCR-ABL1-like ALL is activated kinase signaling, with 50% containing CRLF2 genomic alterations [95] and half of those cases containing concomitant JAK mutations.[96] Additional information about BCR-ABL1-like ALL cases with CRLF2 genomic alterations is provided below. Many of the remaining cases of BCR-ABL1-like ALL have been noted to have a series of translocations with a common theme of involvement of kinases, including ABL1, ABL2, CSF1R, JAK2, and PDGFRB.[4,93] Fusion proteins from these gene combinations have been noted in some cases to be transformative and have responded to tyrosine kinase inhibitors both in vitro and in vivo,[93] suggesting potential therapeutic strategies for these patients. Point mutations in kinase genes, aside from those in JAK1 and JAK2, are uncommon in Ph-like ALL cases.[9] Genomic alterations in CRLF2, a cytokine receptor gene located on the pseudoautosomal regions of the sex chromosomes, have been identified in 5% to 10% of cases of precursor B-cell ALL; they represent approximately 50% of cases of BCR-ABL1-like ALL.[97,98] The chromosomal abnormalities that commonly lead to CRLF2 overexpression include translocations of the IgH locus (chromosome 14) to CRLF2 and interstitial deletions in pseudoautosomal regions of the sex chromosomes, resulting in a P2RY8-CRLF2 fusion.[9,95,97,98]CRLF2 abnormalities are strongly associated with the presence of IKZF1 deletions and JAK mutations;[9,82,95,96,98] they are also more common in children with Down syndrome.[98] Point mutations in tyrosine kinase genes other than JAK1 and JAK2 are uncommon in CRLF2-overexpressing cases.[9] Although the results of several retrospective studies suggest that CRLF2 abnormalities may have adverse prognostic significance on univariate analyses, most do not find this abnormality to be an independent predictor of outcome.[95,97,98,99,100] For example, in a large European study, increased expression of CRLF2 was not associated with unfavorable outcome in multivariate analysis, while IKZF1 deletion and BCR-ABL1-like expression signatures were associated with unfavorable outcome.[89] Controversy exists about whether the prognostic significance of CRLF2 abnormalities should be analyzed based on CRLF2 overexpression or on the presence of CRLF2 genomic alterations.[99,100] Approximately 9% of BCR-ABL1-like ALL cases result from rearrangements that lead to overexpression of a truncated erythropoietin receptor (EPOR).[101] The C-terminal region of the receptor that is lost is the region that is mutated in primary familial congenital polycythemia and that controls stability of the EPOR. The portion of the EPOR remaining is sufficient for JAK-STAT activation and for driving leukemia development.
- Gene polymorphisms in drug metabolic pathways
A number of polymorphisms of genes involved in the metabolism of chemotherapeutic agents have been reported to have prognostic significance in childhood ALL.[102,103,104] For example, patients with mutant phenotypes of thiopurine methyltransferase (TPMT, a gene involved in the metabolism of thiopurines, such as mercaptopurine [6-MP]), appear to have more favorable outcomes,[105] although such patients may also be at higher risk of developing significant treatment-related toxicities, including myelosuppression and infection.[106,107] Patients with homozygosity for TPMT variants associated with low enzymatic activity tolerate only very low doses of mercaptopurine (approximately 10% of the standard dose) and are treated with reduced doses of mercaptopurine to avoid excessive toxicity. Patients who are heterozygous for this mutant enzyme gene generally tolerate mercaptopurine without serious toxicity, but they do require more frequent dose reductions for hematologic toxicity than do patients who are homozygous for the normal allele.[108,109] Germline variants in nucleoside diphosphate-linked moiety X-type motif 15 (NUDT15) that reduce or abolish activity of this enzyme also lead to diminished tolerance to thiopurines.[108,110] The variants are most common in East Asians and Hispanics, and they are rare in Europeans and Africans. Patients homozygous for the risk variants tolerate only very low doses of mercaptopurine, while patients heterozygous for the risk alleles tolerate lower doses than do patients homozygous for the wild-type allele (approximately 25% dose reduction on average), but there is broad overlap in tolerated doses between the two groups.[108,111] Gene polymorphisms may also affect the expression of proteins that play central roles in the cellular effects of anticancer drugs. As an example, patients who are homozygous for a polymorphism in the promoter region of CEP72 (a centrosomal protein involved in microtubule formation) are at increased risk of vincristine neurotoxicity.[112] Genome-wide polymorphism analysis has identified specific single nucleotide polymorphisms associated with high end-induction MRD and risk of relapse. Polymorphisms of IL-15, as well as genes associated with the metabolism of etoposide and methotrexate, were significantly associated with treatment response in two large cohorts of ALL patients treated on SJCRH and COG protocols.[113] Polymorphic variants involving the reduced folate carrier and methotrexate metabolism have been linked to toxicity and outcome.[114,115] While these associations suggest that individual variations in drug metabolism can affect outcome, few studies have attempted to adjust for these variations; it is unknown whether individualized dose modification based on these findings will improve outcome.
(Refer to the PDQ summary on Childhood Acute Lymphoblastic Leukemia Treatment for information about the treatment of childhood ALL.) Acute Myeloid Leukemia (AML) Pediatric AML is typically a disease of recurring chromosomal alterations, with conventional cytogenetics detecting structural and numerical cytogenetic abnormalities in 70% to 80% of children with AML, while the recently recognized cryptic translocations (e.g., NUP98/NSD1, CBFA2T3/GLIS2, and NUP98/KDM5A) and mutations (e.g., CEBPA and NPM1) account for many of the remaining cases.[116,117] Comprehensive molecular profiling of AML in pediatric and adult cases has characterized AML as a disease showing both commonalities and distinct differences between the age groups.[117,118,119]Figure 3 (A) illustrates the frequencies of recurring gene mutations in adult and pediatric AML, showing that some mutations are differentially present between pediatric and adults cases (e.g., IDH1 and DNMT3A mutations being much more common in adults than children).[117]Figure 3 (B) shows that common genomic alterations in adult AML (FLT3-ITD, NPM1, and CEBPA mutations) are uncommon in children younger than 5 years but increase in frequency with age.[117]
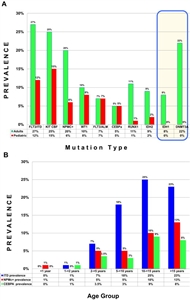
Figure 3. (A) Prevalence of AML-associated mutations in pediatric versus adult AML, demonstrating lower incidence of mutations in pediatric AML. Bordered panel shows 2 newly discovered mutations in adults that are absent in pediatric AML. (B) Age-based prevalence of common AML-associated mutations. Reprinted from Pediatric Clinics of North America, Volume 62, Katherine Tarlock, Soheil Meshinchi, Pediatric Acute Myeloid Leukemia: Biology and Therapeutic Implications of Genomic Variants, Pages 75-93, Copyright (2015), with permission from Elsevier. Figure 4 (A) shows the marked variation in MLL (KMT2A)-rearranged AML by age, with much higher frequencies for infants compared with older children and adults.[117] Normal karyotype AML and core-binding factor AML show an opposing pattern, with very low rates in infancy and with increasing rates in the first two decades of life. Figure 4 (B) shows specific cryptic translocations that occur primarily in children (NUP98/NSD1, CBFA2T3/GLIS2, and NUP98/KDM5A) and vary by age.[117]
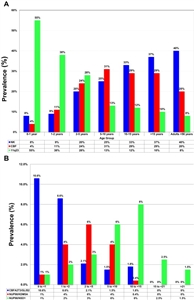
Figure 4. Age-based prevalence of specific karyotypic (A) or cryptic (B) translocations in AML. Reprinted from Pediatric Clinics of North America, Volume 62, Katherine Tarlock, Soheil Meshinchi, Pediatric Acute Myeloid Leukemia: Biology and Therapeutic Implications of Genomic Variants, Pages 75-93, Copyright (2015), with permission from Elsevier. The genomic landscape of pediatric AML cases can change from diagnosis to relapse, with mutations detectable at diagnosis dropping out at relapse and conversely with new mutations appearing at relapse. A key finding in a study of 20 cases for which sequencing data were available at diagnosis and relapse was that the variant allele frequency at diagnosis strongly correlated with persistence of mutations at relapse.[120] Approximately 90% of the diagnostic variants with variant allele frequency greater than 0.4 persisted to relapse compared with only 28% with variant allele frequency less than 0.2 (P < .001). This observation is consistent with previous results showing that presence of the FLT3-ITD mutation predicted for poor prognosis only when there was a high FLT3-ITD allelic ratio. Chromosomal analyses of leukemia (using either conventional cytogenetic methods and/or molecular methods) should be performed on children with AML because chromosomal abnormalities are important diagnostic and prognostic markers.[121,122,123,124,125,126] Clonal chromosomal abnormalities have been identified in the blasts of about 75% of children with AML and are useful in defining subtypes with particular characteristics (e.g., t(8;21), t(15;17), inv(16), 11q23 abnormalities, t(1;22)). Leukemias with the chromosomal abnormalities t(8;21) and inv(16) are called core-binding factor leukemias; core-binding factor (a transcription factor involved in hematopoietic stem cell differentiation) is disrupted by each of these abnormalities. A unifying concept for the role of specific mutations in AML is that mutations that promote proliferation (Type I) and mutations that block normal myeloid development (Type II) are required for full conversion of hematopoietic stem/precursor cells to malignancy.[127,128] Support for this conceptual construct comes from the observation that there is generally mutual exclusivity within each type of mutation, such that a single Type I and a single Type II mutation are present within each case. Further support comes from genetically engineered models of AML for which cooperative events rather than single mutations are required for leukemia development. Type I mutations are commonly in genes involved in growth factor signal transduction and include mutations in FLT3, KIT, NRAS, KRAS, and PTNP11.[129] Type II genomic alterations include the common translocations and mutations associated with favorable prognosis (t(8;21), inv(16), t(16;16), t(15;17), CEBPA, and NPM1). MLL (KMT2A) rearrangements (translocations and partial tandem duplication) are also classified as Type II mutations. Specific recurring cytogenetic and molecular abnormalities are briefly described below. The abnormalities are listed by those in clinical use that identify patients with favorable or unfavorable prognosis, followed by other abnormalities. Molecular abnormalities associated with favorable prognosis include the following:
- t(8;21) (RUNX1-RUNX1T1): In leukemias with t(8;21), the RUNX1(AML1) gene on chromosome 21 is fused with the RUNX1T1 (ETO) gene on chromosome 8. The t(8;21) translocation is associated with the FAB M2 subtype and with granulocytic sarcomas.[130,131] Adults with t(8;21) have a more favorable prognosis than adults with other types of AML.[121,132] These children have a more favorable outcome compared with children with AML characterized by normal or complex karyotypes [121,133,134,135] with 5-year overall survival (OS) of 80% to 90%.[124,125] The t(8;21) translocation occurs in approximately 12% of children with AML.[124,125]
Although both RUNX1-RUNX1T1 and CBFB-MYH11 fusion genes disrupt the activity of core-binding factor, which contains RUNX1 and CBFB, cases with these genomic alterations have distinctive secondary mutations.[136,137] Both subtypes commonly show mutations in genes that activate receptor tyrosine kinase signaling (e.g., NRAS, FLT3 and KIT); NRAS and KIT are the most commonly mutated genes for both subtypes. RUNX1-RUNX1T1 cases additionally have frequent mutations in genes regulating chromatin conformation (e.g., ASXL1 and ASXL2) and genes encoding members of the cohesin complex (approximately 40% and 20% of cases, respectively). Mutations in ASXL1 and ASXL2 and mutations in members of the cohesin complex are rare in CBFB-MYH11 leukemias.[136,137] Data exist (primarily from adults) that these secondary mutations may have prognostic significance,[136,137,138] but further study is required to understand their prognostic significance in children. Additionally, KIT mutations may indicate increased risk of treatment failure for patients with core-binding factor AML, although the prognostic significance of KIT mutations may be dependent on the mutant-allele ratio (high ratio unfavorable) and/or the specific type of mutation (exon 17 mutations unfavorable).[136,137]KIT exon 17 mutations are enriched in patients with RUNX1-RUNX1T1 fusions when compared with patients with CBFB-MYH11 fusions.[137] - inv(16) (CBFB-MYH11): In leukemias with inv(16), the CBF beta gene (CBFB) at chromosome band 16q22 is fused with the MYH11 gene at chromosome band 16p13. The inv(16) translocation is associated with the FAB M4Eo subtype.[139] Inv(16) confers a favorable prognosis for both adults and children with AML [121,133,134,135] with a 5-year OS of about 85%.[124,125] Inv(16) occurs in 7% to 9% of children with AML.[124,125] As noted above, cases with CBFB-MYH11 and cases with RUNX1-RUNX1T1 have distinctive secondary mutations; CBFB-MYH11 secondary mutations are primarily restricted to genes that activate receptor tyrosine kinase signaling (NRAS, FLT3, and KIT).[136,137]
- t(15;17) (PML-RARA): AML with t(15;17) is invariably associated with APL, a distinct subtype of AML that is treated differently than other types of AML because of its marked sensitivity to the differentiating effects of all-trans retinoic acid. The t(15;17) translocation leads to the production of a fusion protein involving the retinoid acid receptor alpha and PML.[140] Other much less common translocations involving the retinoic acid receptor alpha can also result in APL (e.g., t(11;17)(q23;q21) involving the PLZF gene).[141]
- Identification of cases with the t(11;17)(q23;q21) is important because of their decreased sensitivity to all-trans retinoic acid.[140,141] APL represents about 7% of children with AML.[125,142]
- Nucleophosmin (NPM1) mutations: NPM1 is a protein that has been linked to ribosomal protein assembly and transport as well as being a molecular chaperone involved in preventing protein aggregation in the nucleolus. Immunohistochemical methods can be used to accurately identify patients with NPM1 mutations by the demonstration of cytoplasmic localization of NPM.[143] Mutations in the NPM1 protein that diminish its nuclear localization are primarily associated with a subset of AML with a normal karyotype, absence of CD34 expression,[144] and an improved prognosis in the absence of FLT3-internal tandem duplication (ITD) mutations in adults and younger adults.[144,145,146,147,148,149]
Studies of children with AML suggest a lower rate of occurrence of NPM1 mutations in children compared with adults with normal cytogenetics. NPM1 mutations occur in approximately 8% of pediatric patients with AML and are uncommon in children younger than 2 years.[128,150,151,152]NPM1 mutations are associated with a favorable prognosis in patients with AML characterized by a normal karyotype.[128,151,152] For the pediatric population, conflicting reports have been published regarding the prognostic significance of an NPM1 mutation when a FLT3-ITD mutation is also present, with one study reporting that an NPM1 mutation did not completely abrogate the poor prognosis associated with having a FLT3-ITD mutation,[151,153] but with other studies showing no impact of a FLT3-ITD mutation on the favorable prognosis associated with an NPM1 mutation.[128,152] - CEBPA mutations: Mutations in the CCAAT/Enhancer Binding Protein Alpha gene (CEBPA) occur in a subset of children and adults with cytogenetically normal AML.[154] In adults younger than 60 years, approximately 15% of cytogenetically normal AML cases have mutations in CEBPA.[148] Outcome for adults with AML with CEBPA mutations appears to be relatively favorable and similar to that of patients with core-binding factor leukemias.[148,155] Studies in adults with AML have demonstrated that CEBPA double-mutant, but not single-allele mutant, AML was independently associated with a favorable prognosis.[156,157,158,159]
CEBPA mutations occur in 5% to 8% of children with AML and have been preferentially found in the cytogenetically normal subtype of AML with FAB M1 or M2; 70% to 80% of pediatric patients have double-mutant alleles, which is predictive of a significantly improved survival and similar to the effect observed in adult studies.[160,161] Although both double- and single-mutant alleles of CEBPA were associated with a favorable prognosis in children with AML in one large study,[160] a second study observed inferior outcome for patients with single CEBPA mutations.[161] However, very low numbers of children with single-allele mutants were included in these two studies (only 13 in toto), making a conclusion regarding the prognostic significance of single-allele CEBPA mutations in children premature.[160] In newly diagnosed patients with double-mutant CEBPA AML, in addition to usual family history queries, germline screening should be considered because 5% to 10% of these patients are reported to have a germline CEBPA mutation.[154]
Molecular abnormalities associated with an unfavorable prognosis include the following:
- Chromosomes 5 and 7: Chromosomal abnormalities associated with poor prognosis in adults with AML include those involving chromosome 5 (monosomy 5 and del(5q)) and chromosome 7 (monosomy 7).[121,132,162] These cytogenetic subgroups represent approximately 2% and 4% of pediatric AML cases, respectively, and are also associated with poor prognosis in children.[124,132,162,163,164,165]
In the past, patients with del(7q) were also considered to be at high risk of treatment failure and data from adults with AML support a poor prognosis for both del(7q) and monosomy 7.[126] However, outcome for children with del(7q), but not monosomy 7, appears to be comparable to that of other children with AML.[125,165] The presence of del(7q) does not abrogate the prognostic significance of favorable cytogenetic characteristics (e.g., inv(16) and t(8;21)).[121,165,166] Chromosome 5 and 7 abnormalities appear to lack prognostic significance in AML patients with Down syndrome who are 4 years of age and younger.[167] - Chromosome 3 (inv(3)(q21;q26) or t(3;3)(q21;q26)) and EVI1 overexpression: The inv(3) and t(3;3) abnormalities involving the EVI1 gene located at chromosome 3q26 are associated with poor prognosis in adults with AML,[121,132,168] but are very uncommon in children (<1% of pediatric AML cases).[124,134,169]
- FLT3 mutations: Presence of a FLT3-ITD mutation appears to be associated with poor prognosis in adults with AML,[170] particularly when both alleles are mutated or there is a high ratio of the mutant allele to the normal allele.[171,172]FLT3-ITD mutations also convey a poor prognosis in children with AML.[153,173,174,175,176,177] The frequency of FLT3-ITD mutations in children is lower than that observed in adults, especially for children younger than 10 years, for whom 5% to 10% of cases have the mutation (compared with approximately 30% for adults).[175,176,178] The prevalence of FLT3-ITD is increased in certain genomic subtypes of pediatric AML, including those with the NUP98-NSD1 fusion gene, of which 80% to 90% have FLT3-ITD.[179,180] Approximately 15% of patients with FLT3-ITD have NUP98-NSD1, and patients with both FLT3-ITD and NUP98-NSD1 have a poorer prognosis than do patients with FLT3-ITD and without NUP98-NSD1.[180]
For APL, FLT3-ITD and point mutations occur in 30% to 40% of children and adults.[171,174,175,181,182,183,184] Presence of the FLT3-ITD mutation is strongly associated with the microgranular variant (M3v) of APL and with hyperleukocytosis.[174,183,185,186] It remains unclear whether FLT3 mutations are associated with poorer prognosis in patients with APL who are treated with modern therapy that includes all-trans retinoic acid and arsenic trioxide.[181,182,185,187,188] Activating point mutations of FLT3 have also been identified in both adults and children with AML, though the clinical significance of these mutations is not clearly defined.
Other molecular abnormalities observed in pediatric AML include the following:
- MLL (KMT2A) gene rearrangements: Translocations of chromosomal band 11q23 involving the MLL gene, including most AMLs secondary to epipodophyllotoxin,[189] are associated with monocytic differentiation (FAB M4 and M5). MLL rearrangements are also reported in 5% to 10% of FAB M7 (AMKL) patients.[190] The most common translocation, representing approximately 50% of MLL-rearranged cases in the pediatric AML population, is t(9;11)(p22;q23) in which the MLL gene is fused with the MLLT3(AF9) gene.[191] An MLL gene rearrangement occurs in approximately 20% of children with AML.[124,125] However, more than 50 different fusion partners have been identified for the MLL gene in patients with AML. The median age for 11q23/MLL-rearranged cases in the pediatric AML setting is approximately 2 years, and most translocation subgroups have a median age at presentation of younger than 5 years.[191] However, pediatric cases with t(6;11)(q27;q23) and t(11;17)(q23;q21) have significantly older median ages at presentation (12 years and 9 years, respectively).[191]
Outcome for patients with de novo AML and MLL (KMT2A) gene rearrangement is generally reported as being similar to that for other patients with AML.[121,124,191,192] However, as the MLL gene can participate in translocations with many different fusion partners, the specific fusion partner appears to influence prognosis, as demonstrated by a large international retrospective study evaluating outcome for 756 children with 11q23- or MLL-rearranged AML.[191] For example, cases with t(1;11)(q21;q23), representing 3% of all 11q23/MLL-rearranged AML, showed a highly favorable outcome with 5-year event-free survival (EFS) of 92%. While reports from single clinical trial groups have variably described more favorable prognosis for cases with t(9;11), in which the MLL gene is fused with the AF9 gene, the international retrospective study did not confirm the favorable prognosis of the t(9;11)(p22;q23) subgroup.[121,124,191,193,194,195] An international collaboration evaluating pediatric AMKL observed that the presence of t(9;11), which was seen in approximately 5% of AMKL cases, was associated with an inferior outcome compared with other AMKL cases.[190] Several 11q23/MLL (KMT2A)-rearranged AML subgroups appear to be associated with poor outcome. For example, cases with the t(10;11) translocation are a group at high risk of relapse in bone marrow and the CNS.[121,125,196] Some cases with the t(10;11) translocation have fusion of the MLL gene with the AF10-MLLT10 at 10p12, while others have fusion of MLL with ABI1 at 10p11.2.[197,198] The international retrospective study found that these cases, which present at a median age of approximately 1 year, have a 5-year EFS in the 20% to 30% range.[191] Patients with t(6;11)(q27;q23) and with t(4;11)(q21;q23) also have a poor outcome, with a 5-year EFS of 11% and 29%, respectively, in the international retrospective study.[191] A follow-up study by the international collaborative group demonstrated that additional cytogenetic abnormalities further influenced outcome of children with MLL translocations, with complex karyotypes and trisomy 19 predicting poor outcome and trisomy 8 predicting a more favorable outcome.[199] - t(6;9) (DEK-NUP214): t(6;9) leads to the formation of a leukemia-associated fusion protein DEK-NUP214.[200,201] This subgroup of AML has been associated with a poor prognosis in adults with AML,[200,202,203] and occurs infrequently in children (less than 1% of AML cases). The median age of children with DEK-NUP214 AML is 10 to 11 years, and approximately 40% of pediatric patients have FLT3-ITD.[204,205] t(6;9) AML appears to be associated with a high risk of treatment failure in children, particularly for those not proceeding to allogeneic stem cell transplantation.[124,201,204,205]
- t(1;22) (RBM15-MKL1): The t(1;22)(p13;q13) translocation is uncommon (<1% of pediatric AML) and is restricted to acute megakaryocytic leukemia (AMKL).[124,206,207,208] Studies have found that t(1;22)(p13;q13) is observed in 12% to 14% of children with AMKL and evaluable cytogenetics [190] or molecular genetics.[209] Most AMKL cases with t(1;22) occur in infants, with the median age at presentation (4-7 months) being younger than that for other children with AMKL.[190,210,211] The translocation is uncommon in children with Down syndrome who develop AMKL.[206,208] In leukemias with t(1;22), the RBM15 (OTT) gene on chromosome 1 is fused to the MKL1 (MAL) gene on chromosome 22.[212,213] Cases with detectable RBM15-MKL1 fusion transcripts in the absence of t(1;22) have also been reported.[208]
Controversy exists regarding the prognostic significance of the t(1;22) in pediatric AMKL. In a report from the Berlin-Frankfurt-Münster (BFM) study group of 97 non-Down syndrome AMKL patients, presence of t(1;22) (n = 8) was associated with a significantly inferior outcome (5-year EFS, 38% vs. 53% in other AMKL patients), although all of the observed events in patients with t(1;22) were related to treatment-related mortality.[214] An international collaborative retrospective study with a larger number of t(1;22) cases reported that patients with this abnormality had a 5-year EFS of 54.5% and an OS of 58.2%, similar to the rates for other children with AMKL.[190] In another international retrospective analysis of 153 cases with non-Down syndrome AMKL with samples available for molecular analysis, the 4-year EFS for patients with t(1;22) was 59% and OS was 70%, significantly better than AMKL patients with other specific genetic abnormalities (CBFA2T3/GUS2, NUP98/KDM5A, KMT2A rearrangements, monosomy7).[209] - t(8;16) (MYST3-CREBBP): The t(8;16) translocation fuses the MYST3 gene on chromosome 8p11 to CREBBP on chromosome 16p13. t(8;16) AML occurs rarely in children, and in an international BFM AML study of 62 children, presence of this translocation was associated with younger age at diagnosis (median, 1.2 years), FAB M4/M5 phenotype, erythrophagocytosis, leukemia cutis, and disseminated intravascular coagulation.[215] Outcome for children with t(8;16) AML appears similar to other types of AML. A substantial proportion of infants diagnosed with t(8;16) AML in the first month of life show spontaneous remission, although AML recurrence may occur months to years later.[215,216,217,218,219,220,221] These observations suggest that a watch and wait policy could be considered in cases of t(8;16) AML diagnosed in the neonatal period if close long-term monitoring can be ensured.[215]
- t(7;12)(q36;p13): The t(7;12)(q36;p13) translocation involves ETV6 on chromosome 12p13 and variable breakpoints on chromosome 7q36 in the region of MNX1 (HLXB9).[222] The translocation may be cryptic by conventional karyotyping and in some cases may be confirmed only by FISH.[223,224,225] This alteration occurs virtually exclusively in children younger than 2 years, is mutually exclusive with MLL (KMT2A) rearrangement, and is associated with a high risk of treatment failure.[124,125,128,223,224,226]
- NUP98 gene fusions: NUP98 has been reported to form leukemogenic gene fusions with more than 20 different partners.[227] In the pediatric AML setting, the two most common fusion genes are NUP98-NSD1 and NUP98-JARID1A, with the former observed in one report in approximately 15% of cytogenetically normal pediatric AML and the latter observed in approximately 10% of pediatric AMKL.[179,210] AML cases with either NUP98 fusion gene show high expression of HOXA and HOXB genes, indicative of a stem cell phenotype.[201,210]
- NUP98-NSD1: The NUP98-NSD1 fusion gene, which is often cytogenetically cryptic, results from the fusion of NUP98 (chromosome 11p15) with NSD1 (chromosome 5q35).[179,180,201,228,229,230,231] This alteration occurs in approximately 4% to 5% of pediatric AML cases.[179,201,230] The highest frequency in the pediatric population is in the 5- to 9-year age group (approximately 8%), with lower frequency in younger children (approximately 2% in children younger than 2 years). NUP98-NSD1 cases present with high WBC count (median, 147 × 109 /L in one study).[179,180] Most NUP98-NSD1 AML cases do not show cytogenetic aberrations.[179,201,228] A high percentage of NUP98-NSD1 cases (80% to 90%) have FLT3-ITD.[179,180] A study that included 12 children with NUP98-NSD1 AML reported that although all patients achieved CR, presence of NUP98-NSD1 independently predicted for poor prognosis, and children with NUP98-NSD1 AML had a high risk of relapse, with a resulting 4-year EFS of approximately 10%.[179] In another study that included children (n = 38) and adults (n = 7) with NUP98-NSD1 AML, presence of both NUP98-NSD1 and FLT3-ITD independently predicted for poor prognosis; patients with both lesions had a low CR rate (approximately 30%) and a low 3-year EFS rate (approximately 15%).[180]
- NUP98-JARID1A (also called NUP98/KDM5A): NUP98-JARID1A is a recurrent cryptic translocation in pediatric AMKL, accounting for 9% to 10% of AMKL cases and having a median age at presentation of approximately 2 years. This lesion appears to confer a high risk of relapse (36% ± 14%) and poor EFS and OS (36% ± 13% for each).[209,210]
- CBFA2T3-GLIS2: CBFA2T3-GLIS2 is a fusion resulting from a cryptic chromosome 16 inversion (inv(16)(p13.3q24.3)) [232,233] that is present in approximately 2% of pediatric AML.[210,232,234,235] It occurs most frequently in non-Down syndrome AMKL (~15% of patients),[210] but also has been observed in other cytogenetically normal pediatric AML subtypes (~4% of patients).[234] It has been associated with an inferior outcome.[209,232,235]
- RAS mutations: Although mutations in RAS have been identified in 20% to 25% of patients with AML, the prognostic significance of these mutations has not been clearly shown.[128,236,237,238] Mutations in NRAS are observed more commonly than KRAS mutations in pediatric AML cases.[128,129]RAS mutations occur with similar frequency for all Type II alteration subtypes with the exception of APL, for which RAS mutations are seldom observed.[128]
- KIT mutations: Mutations in KIT occur in approximately 5% of AML, but in 10% to 40% of AML with core-binding factor abnormalities.[128,129,239,240] The presence of activating KIT mutations in adults with this AML subtype appears to be associated with a poorer prognosis compared with core-binding factor AML without KIT mutation.[239,241,242] The prognostic significance of KIT mutations occurring in pediatric core-binding factor AML remains unclear,[243,244,245,246] although the largest pediatric study reported to date observed no prognostic significance for KIT mutations.[247]
- GATA1 mutations: GATA1 mutations are present in most, if not all, Down syndrome children with either transient myeloproliferative disease or AMKL.[248,249,250,251]GATA1 mutations are not observed in non-Down syndrome children with AMKL or in Down syndrome children with other types of leukemia.[250,251]GATA1 is a transcription factor that is required for normal development of erythroid cells, megakaryocytes, eosinophils, and mast cells.[252]GATA1 mutations confer increased sensitivity to cytarabine by down-regulating cytidine deaminase expression, possibly providing an explanation for the superior outcome of children with Down syndrome and M7 AML when treated with cytarabine-containing regimens.[253]
- WT1 mutations: WT1, a zinc-finger protein regulating gene transcription, is mutated in approximately 10% of cytogenetically normal cases of AML in adults.[254,255,256,257] The WT1 mutation has been shown in some,[254,255,257] but not all,[256] studies to be an independent predictor of worse disease-free, event-free, and OS of adults. In children with AML, WT1 mutations are observed in approximately 10% of cases.[258,259] Cases with WT1 mutations are enriched among children with normal cytogenetics and FLT3-ITD, but are less common among children younger than 3 years.[258,259] AML cases with NUP98-NSD1 are enriched for both FLT3-ITD and WT1 mutations.[179] In univariate analyses, WT1 mutations are predictive of poorer outcome in pediatric patients, but the independent prognostic significance of WT1 mutation status is unclear because of its strong association with FLT3-ITD and its association with NUP98-NSD1.[179,258,259] The largest study of WT1 mutations in children with AML observed that children with WT1 mutations in the absence of FLT3-ITD had outcomes similar to that of children without WT1 mutations, while children with both WT1 mutation and FLT3-ITD had survival rates less than 20%.[258]
- DNMT3A mutations: Mutations of the DNA cytosine methyltransferase gene (DNMT3A) have been identified in approximately 20% of adult AML patients, being virtually absent in patients with favorable cytogenetics but occurring in one-third of adult patients with intermediate-risk cytogenetics.[260] Mutations in this gene are independently associated with poor outcome.[260,261,262]DNMT3A mutations appear to be very uncommon in children.[119]
- IDH1 and IDH2 mutations: Mutations in IDH1 and IDH2, which code for isocitrate dehydrogenase, occur in approximately 20% of adults with AML,[263,264,265,266,267] and they are enriched in patients with NPM1 mutations.[264,265,268] The specific mutations that occur in IDH1 and IDH2 create a novel enzymatic activity that promotes conversion of alpha-ketoglutarate to 2-hydroxyglutarate.[269,270] This novel activity appears to induce a DNA hypermethylation phenotype similar to that observed in AML cases with loss of function mutations in TET2.[268] Mutations in IDH1 and IDH2 are uncommon in pediatric AML, occurring in 0% to 4% of cases.[119,271,272,273,274,275] There is no indication of a negative prognostic effect for IDH1 and IDH2 mutations in children with AML.[271]
- CSF3R mutations: CSF3R is the gene encoding the granulocyte colony-stimulating factor (G-CSF) receptor, and activating mutations in CSF3R are observed in 2% to 3% of pediatric AML cases.[276] These mutations lead to enhanced signaling through the G-CSF receptor, and they are primarily observed in AML with either CEBPA mutations or with core-binding factor abnormalities (RUNX1/RUNX1T1 and inversion 16).[276] The clinical characteristics of and prognosis for patients with CSF3R mutations do not seem to be significantly different from those of patients without CSF3R mutations.
Activating mutations in CSF3R are also observed in patients with severe congenital neutropenia. These mutations are not the cause of severe congenital neutropenia, but rather arise as somatic mutations and can represent an early step in the pathway to AML.[277] In one study of patients with severe congenital neutropenia, 34% of patients who had not developed a myeloid malignancy had CSF3R mutations detectable in peripheral blood neutrophils and mononuclear cells, while 78% of patients who had developed a myeloid malignancy showed CSF3R mutations.[277] A study of 31 patients with severe congenital neutropenia who developed AML or MDS observed CSF3R mutations in approximately 80%, and also observed a high frequency of RUNX1 mutations (approximately 60%), suggesting cooperation between CSF3R and RUNX1 mutations for leukemia development within the context of severe congenital neutropenia.[278] - miR-155 expression: miR-155 is a microRNA that is normally upregulated in hematopoietic cells and myeloid progenitor cells as part of an inflammatory response but when aberrantly dysregulated and highly expressed, it independently enhances survival and growth factor independence through repression of PU.1.[279] A study of 363 adults with cytogenetically normal AML found that miR-155 was highly associated with induction failure, disease-free survival, and OS.[280] An independent study of children with cytogenetically normal AML (N = 198) similarly found miR-155 to be an adverse factor. Induction failure (54% vs. 17%; P < .001), 3-year OS (51% vs. 75%; P = .002), and EFS (32% vs. 59%; P < .001) were all worse in patients with high miR-155 expression. As with adults, children in this trial who had high miR-155 expression were more likely to have FLT3-ITD mutations (69%; P < .001). Multivariate analyses found that miR-155 maintained an independent adverse impact on these outcome parameters when controlling for age; white blood cell count; and FLT3-ITD, CEBPA, and NPM1 mutations.[281]
(Refer to the PDQ summary on Childhood Acute Myeloid Leukemia/Other Myeloid Malignancies Treatment for information about the treatment of childhood AML.) Juvenile Myelomonocytic Leukemia (JMML) The genomic landscape of JMML is characterized by mutations in one of five genes of the Ras pathway:[282,283]NF1, NRAS, KRAS, PTPN11, and CBL. In a series of 118 consecutively diagnosed JMML cases with Ras pathway-activating mutations, PTPN11 was the most commonly mutated gene, accounting for 51% of cases (19% germline and 32% somatic) (refer to Figure 5).[282] Patients with mutated NRAS accounted for 19% of cases, and patients with mutated KRAS accounted for 15% of cases. NF1 and CBL mutations accounted for 8% and 11% of cases, respectively. Although mutations among these five genes are generally mutually exclusive, 10% to 17% of cases have mutations in two of these Ras pathway genes,[282,283] a finding that is associated with poorer prognosis.[282] The mutation rate in JMML leukemia cells is very low, but additional mutations beyond those of the five Ras pathway genes described above are observed.[282,283] Secondary genomic alterations are observed for genes of the transcriptional repressor complex PRC2 (e.g., ASXL1 was mutated in 7%-8% of cases). Some genes associated with myeloproliferative neoplasms in adults are also mutated at low rates in JMML (e.g., SETBP1 was mutated in 7%-9% of cases).[282,283,284]JAK3 mutations are also observed in a small percentage (4%-12%) of JMML cases.[282,283,284] Cases with germline PTPN11 and germline CBL mutations showed low rates of additional mutations (refer to Figure 5).[282]
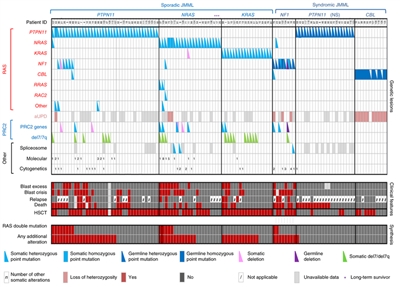
Figure 5. Alteration profiles in individual JMML cases. Germline and somatically acquired alterations with recurring hits in the RAS pathway and PRC2 network are shown for 118 patients with JMML who underwent detailed genetic analysis. Blast excess was defined as a blast count ≥10% but <20% of nucleated cells in the bone marrow at diagnosis. Blast crisis was defined as a blast count ≥20% of nucleated cells in the bone marrow. NS, Noonan syndrome. Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics (Caye A, Strullu M, Guidez F, et al.: Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat Genet 47 [11]: 1334-40, 2015), copyright (2015).
Clinical implications General characteristics of leukemia cells provide both prognostic information and guidance regarding therapeutic opportunities for JMML: - Number of non-RAS pathway mutations. A strong predictor of prognosis for children with JMML is the number of mutations beyond the disease-defining RAS-pathway mutations.[282,283] Of 64 patients (65.3%) at diagnosis, zero or one somatic alteration (pathogenic mutation or monosomy 7) was identified, whereas two or more alterations were identified in 34 (34.7%) patients.[283] In multivariate analysis, mutation number (two or more vs. zero or one) maintained significance as a predictor of inferior event-free survival and overall survival. A higher proportion of patients diagnosed with two or more alterations were older and male, and these patients also demonstrated a higher rate of monosomy 7 or somatic NF1 mutation.[283] Similar findings and observations reported that patients with RAS-pathway double mutations (15 of 96 patients) were at the highest risk of treatment failure.[282]
- RAS-MAPK pathway inhibitors. Because JMML is a disease defined by mutations in the RAS-MAPK pathway, one might speculate that inhibitors of this pathway (e.g., MEK inhibitors) may have clinical utility in the treatment of JMML. However, preclinical data to support this hypothesis are inconsistent,[285,286] and there are no clinical data available.
References:
-
Mullighan CG: Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol 50 (4): 314-24, 2013.
-
Mullighan CG, Goorha S, Radtke I, et al.: Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446 (7137): 758-64, 2007.
-
Mullighan CG, Miller CB, Radtke I, et al.: BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453 (7191): 110-4, 2008.
-
Roberts KG, Li Y, Payne-Turner D, et al.: Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 371 (11): 1005-15, 2014.
-
Harvey RC, Mullighan CG, Wang X, et al.: Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116 (23): 4874-84, 2010.
-
Clappier E, Auclerc MF, Rapion J, et al.: An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia 28 (1): 70-7, 2014.
-
Zaliova M, Zimmermannova O, Dörge P, et al.: ERG deletion is associated with CD2 and attenuates the negative impact of IKZF1 deletion in childhood acute lymphoblastic leukemia. Leukemia 28 (1): 182-5, 2014.
-
Holmfeldt L, Wei L, Diaz-Flores E, et al.: The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 45 (3): 242-52, 2013.
-
Loh ML, Zhang J, Harvey RC, et al.: Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: a report from the Children's Oncology Group TARGET Project. Blood 121 (3): 485-8, 2013.
-
Bercovich D, Ganmore I, Scott LM, et al.: Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet 372 (9648): 1484-92, 2008.
-
Andersson AK, Ma J, Wang J, et al.: The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet 47 (4): 330-7, 2015.
-
Ma X, Edmonson M, Yergeau D, et al.: Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 6: 6604, 2015.
-
Meyer JA, Wang J, Hogan LE, et al.: Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet 45 (3): 290-4, 2013.
-
Li B, Li H, Bai Y, et al.: Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med 21 (6): 563-71, 2015.
-
Mullighan CG, Zhang J, Kasper LH, et al.: CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471 (7337): 235-9, 2011.
-
Moorman AV, Ensor HM, Richards SM, et al.: Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol 11 (5): 429-38, 2010.
-
Arber DA, Orazi A, Hasserjian R, et al.: The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127 (20): 2391-405, 2016.
-
Paulsson K, Johansson B: High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 48 (8): 637-60, 2009.
-
Aricò M, Valsecchi MG, Rizzari C, et al.: Long-term results of the AIEOP-ALL-95 Trial for Childhood Acute Lymphoblastic Leukemia: insight on the prognostic value of DNA index in the framework of Berlin-Frankfurt-Muenster based chemotherapy. J Clin Oncol 26 (2): 283-9, 2008.
-
Dastugue N, Suciu S, Plat G, et al.: Hyperdiploidy with 58-66 chromosomes in childhood B-acute lymphoblastic leukemia is highly curable: 58951 CLG-EORTC results. Blood 121 (13): 2415-23, 2013.
-
Synold TW, Relling MV, Boyett JM, et al.: Blast cell methotrexate-polyglutamate accumulation in vivo differs by lineage, ploidy, and methotrexate dose in acute lymphoblastic leukemia. J Clin Invest 94 (5): 1996-2001, 1994.
-
Moorman AV, Richards SM, Martineau M, et al.: Outcome heterogeneity in childhood high-hyperdiploid acute lymphoblastic leukemia. Blood 102 (8): 2756-62, 2003.
-
Chilton L, Buck G, Harrison CJ, et al.: High hyperdiploidy among adolescents and adults with acute lymphoblastic leukaemia (ALL): cytogenetic features, clinical characteristics and outcome. Leukemia 28 (7): 1511-8, 2014.
-
Sutcliffe MJ, Shuster JJ, Sather HN, et al.: High concordance from independent studies by the Children's Cancer Group (CCG) and Pediatric Oncology Group (POG) associating favorable prognosis with combined trisomies 4, 10, and 17 in children with NCI Standard-Risk B-precursor Acute Lymphoblastic Leukemia: a Children's Oncology Group (COG) initiative. Leukemia 19 (5): 734-40, 2005.
-
Harris MB, Shuster JJ, Carroll A, et al.: Trisomy of leukemic cell chromosomes 4 and 10 identifies children with B-progenitor cell acute lymphoblastic leukemia with a very low risk of treatment failure: a Pediatric Oncology Group study. Blood 79 (12): 3316-24, 1992.
-
Heerema NA, Harbott J, Galimberti S, et al.: Secondary cytogenetic aberrations in childhood Philadelphia chromosome positive acute lymphoblastic leukemia are nonrandom and may be associated with outcome. Leukemia 18 (4): 693-702, 2004.
-
Nachman JB, Heerema NA, Sather H, et al.: Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood 110 (4): 1112-5, 2007.
-
Raimondi SC, Zhou Y, Shurtleff SA, et al.: Near-triploidy and near-tetraploidy in childhood acute lymphoblastic leukemia: association with B-lineage blast cells carrying the ETV6-RUNX1 fusion, T-lineage immunophenotype, and favorable outcome. Cancer Genet Cytogenet 169 (1): 50-7, 2006.
-
Attarbaschi A, Mann G, König M, et al.: Incidence and relevance of secondary chromosome abnormalities in childhood TEL/AML1+ acute lymphoblastic leukemia: an interphase FISH analysis. Leukemia 18 (10): 1611-6, 2004.
-
Lemez P, Attarbaschi A, Béné MC, et al.: Childhood near-tetraploid acute lymphoblastic leukemia: an EGIL study on 36 cases. Eur J Haematol 85 (4): 300-8, 2010.
-
Paulsson K, Lilljebjörn H, Biloglav A, et al.: The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet 47 (6): 672-6, 2015.
-
Harrison CJ, Moorman AV, Broadfield ZJ, et al.: Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br J Haematol 125 (5): 552-9, 2004.
-
Mullighan CG, Jeha S, Pei D, et al.: Outcome of children with hypodiploid ALL treated with risk-directed therapy based on MRD levels. Blood 126 (26): 2896-9, 2015.
-
Irving J, Matheson E, Minto L, et al.: Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 124 (23): 3420-30, 2014.
-
Rubnitz JE, Wichlan D, Devidas M, et al.: Prospective analysis of TEL gene rearrangements in childhood acute lymphoblastic leukemia: a Children's Oncology Group study. J Clin Oncol 26 (13): 2186-91, 2008.
-
Kanerva J, Saarinen-Pihkala UM, Niini T, et al.: Favorable outcome in 20-year follow-up of children with very-low-risk ALL and minimal standard therapy, with special reference to TEL-AML1 fusion. Pediatr Blood Cancer 42 (1): 30-5, 2004.
-
Aldrich MC, Zhang L, Wiemels JL, et al.: Cytogenetics of Hispanic and White children with acute lymphoblastic leukemia in California. Cancer Epidemiol Biomarkers Prev 15 (3): 578-81, 2006.
-
Loh ML, Goldwasser MA, Silverman LB, et al.: Prospective analysis of TEL/AML1-positive patients treated on Dana-Farber Cancer Institute Consortium Protocol 95-01. Blood 107 (11): 4508-13, 2006.
-
Borowitz MJ, Devidas M, Hunger SP, et al.: Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children's Oncology Group study. Blood 111 (12): 5477-85, 2008.
-
Madzo J, Zuna J, Muzíková K, et al.: Slower molecular response to treatment predicts poor outcome in patients with TEL/AML1 positive acute lymphoblastic leukemia: prospective real-time quantitative reverse transcriptase-polymerase chain reaction study. Cancer 97 (1): 105-13, 2003.
-
Bhojwani D, Pei D, Sandlund JT, et al.: ETV6-RUNX1-positive childhood acute lymphoblastic leukemia: improved outcome with contemporary therapy. Leukemia 26 (2): 265-70, 2012.
-
Enshaei A, Schwab CJ, Konn ZJ, et al.: Long-term follow-up of ETV6-RUNX1 ALL reveals that NCI risk, rather than secondary genetic abnormalities, is the key risk factor. Leukemia 27 (11): 2256-9, 2013.
-
Barbany G, Andersen MK, Autio K, et al.: Additional aberrations of the ETV6 and RUNX1 genes have no prognostic impact in 229 t(12;21)(p13;q22)-positive B-cell precursor acute lymphoblastic leukaemias treated according to the NOPHO-ALL-2000 protocol. Leuk Res 36 (7): 936-8, 2012.
-
Forestier E, Heyman M, Andersen MK, et al.: Outcome of ETV6/RUNX1-positive childhood acute lymphoblastic leukaemia in the NOPHO-ALL-1992 protocol: frequent late relapses but good overall survival. Br J Haematol 140 (6): 665-72, 2008.
-
Seeger K, Stackelberg AV, Taube T, et al.: Relapse of TEL-AML1--positive acute lymphoblastic leukemia in childhood: a matched-pair analysis. J Clin Oncol 19 (13): 3188-93, 2001.
-
Gandemer V, Chevret S, Petit A, et al.: Excellent prognosis of late relapses of ETV6/RUNX1-positive childhood acute lymphoblastic leukemia: lessons from the FRALLE 93 protocol. Haematologica 97 (11): 1743-50, 2012.
-
Zuna J, Ford AM, Peham M, et al.: TEL deletion analysis supports a novel view of relapse in childhood acute lymphoblastic leukemia. Clin Cancer Res 10 (16): 5355-60, 2004.
-
van Delft FW, Horsley S, Colman S, et al.: Clonal origins of relapse in ETV6-RUNX1 acute lymphoblastic leukemia. Blood 117 (23): 6247-54, 2011.
-
Aricò M, Schrappe M, Hunger SP, et al.: Clinical outcome of children with newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia treated between 1995 and 2005. J Clin Oncol 28 (31): 4755-61, 2010.
-
Schrappe M, Aricò M, Harbott J, et al.: Philadelphia chromosome-positive (Ph+) childhood acute lymphoblastic leukemia: good initial steroid response allows early prediction of a favorable treatment outcome. Blood 92 (8): 2730-41, 1998.
-
Ribeiro RC, Broniscer A, Rivera GK, et al.: Philadelphia chromosome-positive acute lymphoblastic leukemia in children: durable responses to chemotherapy associated with low initial white blood cell counts. Leukemia 11 (9): 1493-6, 1997.
-
Biondi A, Schrappe M, De Lorenzo P, et al.: Imatinib after induction for treatment of children and adolescents with Philadelphia-chromosome-positive acute lymphoblastic leukaemia (EsPhALL): a randomised, open-label, intergroup study. Lancet Oncol 13 (9): 936-45, 2012.
-
Schultz KR, Bowman WP, Aledo A, et al.: Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children's oncology group study. J Clin Oncol 27 (31): 5175-81, 2009.
-
Schultz KR, Carroll A, Heerema NA, et al.: Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children's Oncology Group study AALL0031. Leukemia 28 (7): 1467-71, 2014.
-
Pui CH, Chessells JM, Camitta B, et al.: Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia 17 (4): 700-6, 2003.
-
Johansson B, Moorman AV, Haas OA, et al.: Hematologic malignancies with t(4;11)(q21;q23)--a cytogenetic, morphologic, immunophenotypic and clinical study of 183 cases. European 11q23 Workshop participants. Leukemia 12 (5): 779-87, 1998.
-
Raimondi SC, Peiper SC, Kitchingman GR, et al.: Childhood acute lymphoblastic leukemia with chromosomal breakpoints at 11q23. Blood 73 (6): 1627-34, 1989.
-
Harrison CJ, Moorman AV, Barber KE, et al.: Interphase molecular cytogenetic screening for chromosomal abnormalities of prognostic significance in childhood acute lymphoblastic leukaemia: a UK Cancer Cytogenetics Group Study. Br J Haematol 129 (4): 520-30, 2005.
-
Pui CH, Pei D, Campana D, et al.: A revised definition for cure of childhood acute lymphoblastic leukemia. Leukemia 28 (12): 2336-43, 2014.
-
Pieters R, Schrappe M, De Lorenzo P, et al.: A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet 370 (9583): 240-50, 2007.
-
Pui CH, Gaynon PS, Boyett JM, et al.: Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet 359 (9321): 1909-15, 2002.
-
Rubnitz JE, Camitta BM, Mahmoud H, et al.: Childhood acute lymphoblastic leukemia with the MLL-ENL fusion and t(11;19)(q23;p13.3) translocation. J Clin Oncol 17 (1): 191-6, 1999.
-
Hunger SP: Chromosomal translocations involving the E2A gene in acute lymphoblastic leukemia: clinical features and molecular pathogenesis. Blood 87 (4): 1211-24, 1996.
-
Uckun FM, Sensel MG, Sather HN, et al.: Clinical significance of translocation t(1;19) in childhood acute lymphoblastic leukemia in the context of contemporary therapies: a report from the Children's Cancer Group. J Clin Oncol 16 (2): 527-35, 1998.
-
Fischer U, Forster M, Rinaldi A, et al.: Genomics and drug profiling of fatal TCF3-HLF-positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options. Nat Genet 47 (9): 1020-9, 2015.
-
Pui CH, Sandlund JT, Pei D, et al.: Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA 290 (15): 2001-7, 2003.
-
Crist WM, Carroll AJ, Shuster JJ, et al.: Poor prognosis of children with pre-B acute lymphoblastic leukemia is associated with the t(1;19)(q23;p13): a Pediatric Oncology Group study. Blood 76 (1): 117-22, 1990.
-
Andersen MK, Autio K, Barbany G, et al.: Paediatric B-cell precursor acute lymphoblastic leukaemia with t(1;19)(q23;p13): clinical and cytogenetic characteristics of 47 cases from the Nordic countries treated according to NOPHO protocols. Br J Haematol 155 (2): 235-43, 2011.
-
Pui CH, Campana D, Pei D, et al.: Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 360 (26): 2730-41, 2009.
-
Jeha S, Pei D, Raimondi SC, et al.: Increased risk for CNS relapse in pre-B cell leukemia with the t(1;19)/TCF3-PBX1. Leukemia 23 (8): 1406-9, 2009.
-
Minson KA, Prasad P, Vear S, et al.: t(17;19) in Children with Acute Lymphocytic Leukemia: A Report of 3 Cases and a Review of the Literature. Case Rep Hematol 2013: 563291, 2013.
-
Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, et al.: Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun 7: 11790, 2016.
-
Zhang J, McCastlain K, Yoshihara H, et al.: Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet 48 (12): 1481-1489, 2016.
-
Hogan TF, Koss W, Murgo AJ, et al.: Acute lymphoblastic leukemia with chromosomal 5;14 translocation and hypereosinophilia: case report and literature review. J Clin Oncol 5 (3): 382-90, 1987.
-
Grimaldi JC, Meeker TC: The t(5;14) chromosomal translocation in a case of acute lymphocytic leukemia joins the interleukin-3 gene to the immunoglobulin heavy chain gene. Blood 73 (8): 2081-5, 1989.
-
Meeker TC, Hardy D, Willman C, et al.: Activation of the interleukin-3 gene by chromosome translocation in acute lymphocytic leukemia with eosinophilia. Blood 76 (2): 285-9, 1990.
-
Sutton R, Lonergan M, Tapp H, et al.: Two cases of hypereosinophilia and high-risk acute lymphoblastic leukemia. Leukemia 22 (7): 1463-5, 2008.
-
Heerema NA, Carroll AJ, Devidas M, et al.: Intrachromosomal amplification of chromosome 21 is associated with inferior outcomes in children with acute lymphoblastic leukemia treated in contemporary standard-risk children's oncology group studies: a report from the children's oncology group. J Clin Oncol 31 (27): 3397-402, 2013.
-
Moorman AV, Robinson H, Schwab C, et al.: Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: a comparison of the MRC ALL97/99 and UKALL2003 trials. J Clin Oncol 31 (27): 3389-96, 2013.
-
Harrison CJ, Moorman AV, Schwab C, et al.: An international study of intrachromosomal amplification of chromosome 21 (iAMP21): cytogenetic characterization and outcome. Leukemia 28 (5): 1015-21, 2014.
-
Mullighan CG, Su X, Zhang J, et al.: Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360 (5): 470-80, 2009.
-
Schwab CJ, Chilton L, Morrison H, et al.: Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica 98 (7): 1081-8, 2013.
-
Clappier E, Grardel N, Bakkus M, et al.: IKZF1 deletion is an independent prognostic marker in childhood B-cell precursor acute lymphoblastic leukemia, and distinguishes patients benefiting from pulses during maintenance therapy: results of the EORTC Children's Leukemia Group study 58951. Leukemia 29 (11): 2154-61, 2015.
-
Buitenkamp TD, Pieters R, Gallimore NE, et al.: Outcome in children with Down's syndrome and acute lymphoblastic leukemia: role of IKZF1 deletions and CRLF2 aberrations. Leukemia 26 (10): 2204-11, 2012.
-
Den Boer ML, van Slegtenhorst M, De Menezes RX, et al.: A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol 10 (2): 125-34, 2009.
-
Krentz S, Hof J, Mendioroz A, et al.: Prognostic value of genetic alterations in children with first bone marrow relapse of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia 27 (2): 295-304, 2013.
-
Feng J, Tang Y: Prognostic significance of IKZF1 alteration status in pediatric B-lineage acute lymphoblastic leukemia: a meta-analysis. Leuk Lymphoma 54 (4): 889-91, 2013.
-
Dörge P, Meissner B, Zimmermann M, et al.: IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica 98 (3): 428-32, 2013.
-
van der Veer A, Waanders E, Pieters R, et al.: Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood 122 (15): 2622-9, 2013.
-
Olsson L, Castor A, Behrendtz M, et al.: Deletions of IKZF1 and SPRED1 are associated with poor prognosis in a population-based series of pediatric B-cell precursor acute lymphoblastic leukemia diagnosed between 1992 and 2011. Leukemia 28 (2): 302-10, 2014.
-
Boer JM, van der Veer A, Rizopoulos D, et al.: Prognostic value of rare IKZF1 deletion in childhood B-cell precursor acute lymphoblastic leukemia: an international collaborative study. Leukemia 30 (1): 32-8, 2016.
-
van der Veer A, Zaliova M, Mottadelli F, et al.: IKZF1 status as a prognostic feature in BCR-ABL1-positive childhood ALL. Blood 123 (11): 1691-8, 2014.
-
Roberts KG, Morin RD, Zhang J, et al.: Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 22 (2): 153-66, 2012.
-
Roberts KG, Pei D, Campana D, et al.: Outcomes of children with BCR-ABL1-like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol 32 (27): 3012-20, 2014.
-
Harvey RC, Mullighan CG, Chen IM, et al.: Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood 115 (26): 5312-21, 2010.
-
Mullighan CG, Collins-Underwood JR, Phillips LA, et al.: Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet 41 (11): 1243-6, 2009.
-
Cario G, Zimmermann M, Romey R, et al.: Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood 115 (26): 5393-7, 2010.
-
Ensor HM, Schwab C, Russell LJ, et al.: Demographic, clinical, and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood 117 (7): 2129-36, 2011.
-
Chen IM, Harvey RC, Mullighan CG, et al.: Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children's Oncology Group study. Blood 119 (15): 3512-22, 2012.
-
Palmi C, Vendramini E, Silvestri D, et al.: Poor prognosis for P2RY8-CRLF2 fusion but not for CRLF2 over-expression in children with intermediate risk B-cell precursor acute lymphoblastic leukemia. Leukemia 26 (10): 2245-53, 2012.
-
Iacobucci I, Li Y, Roberts KG, et al.: Truncating Erythropoietin Receptor Rearrangements in Acute Lymphoblastic Leukemia. Cancer Cell 29 (2): 186-200, 2016.
-
Davies SM, Bhatia S, Ross JA, et al.: Glutathione S-transferase genotypes, genetic susceptibility, and outcome of therapy in childhood acute lymphoblastic leukemia. Blood 100 (1): 67-71, 2002.
-
Krajinovic M, Costea I, Chiasson S: Polymorphism of the thymidylate synthase gene and outcome of acute lymphoblastic leukaemia. Lancet 359 (9311): 1033-4, 2002.
-
Krajinovic M, Lemieux-Blanchard E, Chiasson S, et al.: Role of polymorphisms in MTHFR and MTHFD1 genes in the outcome of childhood acute lymphoblastic leukemia. Pharmacogenomics J 4 (1): 66-72, 2004.
-
Schmiegelow K, Forestier E, Kristinsson J, et al.: Thiopurine methyltransferase activity is related to the risk of relapse of childhood acute lymphoblastic leukemia: results from the NOPHO ALL-92 study. Leukemia 23 (3): 557-64, 2009.
-
Relling MV, Hancock ML, Boyett JM, et al.: Prognostic importance of 6-mercaptopurine dose intensity in acute lymphoblastic leukemia. Blood 93 (9): 2817-23, 1999.
-
Stanulla M, Schaeffeler E, Flohr T, et al.: Thiopurine methyltransferase (TPMT) genotype and early treatment response to mercaptopurine in childhood acute lymphoblastic leukemia. JAMA 293 (12): 1485-9, 2005.
-
Yang JJ, Landier W, Yang W, et al.: Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol 33 (11): 1235-42, 2015.
-
Relling MV, Hancock ML, Rivera GK, et al.: Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst 91 (23): 2001-8, 1999.
-
Moriyama T, Nishii R, Perez-Andreu V, et al.: NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet 48 (4): 367-73, 2016.
-
Tanaka Y, Kato M, Hasegawa D, et al.: Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. Br J Haematol 171 (1): 109-15, 2015.
-
Diouf B, Crews KR, Lew G, et al.: Association of an inherited genetic variant with vincristine-related peripheral neuropathy in children with acute lymphoblastic leukemia. JAMA 313 (8): 815-23, 2015.
-
Yang JJ, Cheng C, Yang W, et al.: Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA 301 (4): 393-403, 2009.
-
Gregers J, Christensen IJ, Dalhoff K, et al.: The association of reduced folate carrier 80G>A polymorphism to outcome in childhood acute lymphoblastic leukemia interacts with chromosome 21 copy number. Blood 115 (23): 4671-7, 2010.
-
Radtke S, Zolk O, Renner B, et al.: Germline genetic variations in methotrexate candidate genes are associated with pharmacokinetics, toxicity, and outcome in childhood acute lymphoblastic leukemia. Blood 121 (26): 5145-53, 2013.
-
Creutzig U, van den Heuvel-Eibrink MM, Gibson B, et al.: Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood 120 (16): 3187-205, 2012.
-
Tarlock K, Meshinchi S: Pediatric acute myeloid leukemia: biology and therapeutic implications of genomic variants. Pediatr Clin North Am 62 (1): 75-93, 2015.
-
Schuback HL, Arceci RJ, Meshinchi S: Somatic characterization of pediatric acute myeloid leukemia using next-generation sequencing. Semin Hematol 50 (4): 325-32, 2013.
-
Ho PA, Kutny MA, Alonzo TA, et al.: Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: a report from the Children's Oncology Group. Pediatr Blood Cancer 57 (2): 204-9, 2011.
-
Farrar JE, Schuback HL, Ries RE, et al.: Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Res 76 (8): 2197-205, 2016.
-
Grimwade D, Walker H, Oliver F, et al.: The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood 92 (7): 2322-33, 1998.
-
Gilliland DG: Targeted therapies in myeloid leukemias. Ann Hematol 83 (Suppl 1): S75-6, 2004.
-
Avivi I, Rowe JM: Prognostic factors in acute myeloid leukemia. Curr Opin Hematol 12 (1): 62-7, 2005.
-
Harrison CJ, Hills RK, Moorman AV, et al.: Cytogenetics of childhood acute myeloid leukemia: United Kingdom Medical Research Council Treatment trials AML 10 and 12. J Clin Oncol 28 (16): 2674-81, 2010.
-
von Neuhoff C, Reinhardt D, Sander A, et al.: Prognostic impact of specific chromosomal aberrations in a large group of pediatric patients with acute myeloid leukemia treated uniformly according to trial AML-BFM 98. J Clin Oncol 28 (16): 2682-9, 2010.
-
Grimwade D, Hills RK, Moorman AV, et al.: Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 116 (3): 354-65, 2010.
-
Gilliland DG, Griffin JD: The roles of FLT3 in hematopoiesis and leukemia. Blood 100 (5): 1532-42, 2002.
-
Balgobind BV, Hollink IH, Arentsen-Peters ST, et al.: Integrative analysis of type-I and type-II aberrations underscores the genetic heterogeneity of pediatric acute myeloid leukemia. Haematologica 96 (10): 1478-87, 2011.
-
Kühn MW, Radtke I, Bullinger L, et al.: High-resolution genomic profiling of adult and pediatric core-binding factor acute myeloid leukemia reveals new recurrent genomic alterations. Blood 119 (10): e67-75, 2012.
-
Rubnitz JE, Raimondi SC, Halbert AR, et al.: Characteristics and outcome of t(8;21)-positive childhood acute myeloid leukemia: a single institution's experience. Leukemia 16 (10): 2072-7, 2002.
-
Tallman MS, Hakimian D, Shaw JM, et al.: Granulocytic sarcoma is associated with the 8;21 translocation in acute myeloid leukemia. J Clin Oncol 11 (4): 690-7, 1993.
-
Mrózek K, Heerema NA, Bloomfield CD: Cytogenetics in acute leukemia. Blood Rev 18 (2): 115-36, 2004.
-
Creutzig U, Zimmermann M, Ritter J, et al.: Definition of a standard-risk group in children with AML. Br J Haematol 104 (3): 630-9, 1999.
-
Raimondi SC, Chang MN, Ravindranath Y, et al.: Chromosomal abnormalities in 478 children with acute myeloid leukemia: clinical characteristics and treatment outcome in a cooperative pediatric oncology group study-POG 8821. Blood 94 (11): 3707-16, 1999.
-
Lie SO, Abrahamsson J, Clausen N, et al.: Treatment stratification based on initial in vivo response in acute myeloid leukaemia in children without Down's syndrome: results of NOPHO-AML trials. Br J Haematol 122 (2): 217-25, 2003.
-
Duployez N, Marceau-Renaut A, Boissel N, et al.: Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood 127 (20): 2451-9, 2016.
-
Faber ZJ, Chen X, Gedman AL, et al.: The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet 48 (12): 1551-1556, 2016.
-
Eisfeld AK, Kohlschmidt J, Schwind S, et al.: Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia. Leukemia : , 2017.
-
Larson RA, Williams SF, Le Beau MM, et al.: Acute myelomonocytic leukemia with abnormal eosinophils and inv(16) or t(16;16) has a favorable prognosis. Blood 68 (6): 1242-9, 1986.
-
Mistry AR, Pedersen EW, Solomon E, et al.: The molecular pathogenesis of acute promyelocytic leukaemia: implications for the clinical management of the disease. Blood Rev 17 (2): 71-97, 2003.
-
Licht JD, Chomienne C, Goy A, et al.: Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17). Blood 85 (4): 1083-94, 1995.
-
Smith MA, Ries LA, Gurney JG, et al.: Leukemia. In: Ries LA, Smith MA, Gurney JG, et al., eds.: Cancer incidence and survival among children and adolescents: United States SEER Program 1975-1995. Bethesda, Md: National Cancer Institute, SEER Program, 1999. NIH Pub.No. 99-4649, pp 17-34. Also available online. Last accessed January 27, 2017.
-
Falini B, Martelli MP, Bolli N, et al.: Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood 108 (6): 1999-2005, 2006.
-
Falini B, Mecucci C, Tiacci E, et al.: Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 352 (3): 254-66, 2005.
-
Döhner K, Schlenk RF, Habdank M, et al.: Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood 106 (12): 3740-6, 2005.
-
Verhaak RG, Goudswaard CS, van Putten W, et al.: Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 106 (12): 3747-54, 2005.
-
Schnittger S, Schoch C, Kern W, et al.: Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood 106 (12): 3733-9, 2005.
-
Schlenk RF, Döhner K, Krauter J, et al.: Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 358 (18): 1909-18, 2008.
-
Gale RE, Green C, Allen C, et al.: The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood 111 (5): 2776-84, 2008.
-
Cazzaniga G, Dell'Oro MG, Mecucci C, et al.: Nucleophosmin mutations in childhood acute myelogenous leukemia with normal karyotype. Blood 106 (4): 1419-22, 2005.
-
Brown P, McIntyre E, Rau R, et al.: The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood 110 (3): 979-85, 2007.
-
Hollink IH, Zwaan CM, Zimmermann M, et al.: Favorable prognostic impact of NPM1 gene mutations in childhood acute myeloid leukemia, with emphasis on cytogenetically normal AML. Leukemia 23 (2): 262-70, 2009.
-
Staffas A, Kanduri M, Hovland R, et al.: Presence of FLT3-ITD and high BAALC expression are independent prognostic markers in childhood acute myeloid leukemia. Blood 118 (22): 5905-13, 2011.
-
Tawana K, Wang J, Renneville A, et al.: Disease evolution and outcomes in familial AML with germline CEBPA mutations. Blood 126 (10): 1214-23, 2015.
-
Marcucci G, Maharry K, Radmacher MD, et al.: Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol 26 (31): 5078-87, 2008.
-
Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, et al.: Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 113 (13): 3088-91, 2009.
-
Dufour A, Schneider F, Metzeler KH, et al.: Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol 28 (4): 570-7, 2010.
-
Taskesen E, Bullinger L, Corbacioglu A, et al.: Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood 117 (8): 2469-75, 2011.
-
Fasan A, Haferlach C, Alpermann T, et al.: The role of different genetic subtypes of CEBPA mutated AML. Leukemia 28 (4): 794-803, 2014.
-
Ho PA, Alonzo TA, Gerbing RB, et al.: Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children's Oncology Group. Blood 113 (26): 6558-66, 2009.
-
Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, et al.: Characterization of CEBPA mutations and promoter hypermethylation in pediatric acute myeloid leukemia. Haematologica 96 (3): 384-92, 2011.
-
Johnston DL, Alonzo TA, Gerbing RB, et al.: Outcome of pediatric patients with acute myeloid leukemia (AML) and -5/5q- abnormalities from five pediatric AML treatment protocols: a report from the Children's Oncology Group. Pediatr Blood Cancer 60 (12): 2073-8, 2013.
-
Stevens RF, Hann IM, Wheatley K, et al.: Marked improvements in outcome with chemotherapy alone in paediatric acute myeloid leukemia: results of the United Kingdom Medical Research Council's 10th AML trial. MRC Childhood Leukaemia Working Party. Br J Haematol 101 (1): 130-40, 1998.
-
Wells RJ, Arthur DC, Srivastava A, et al.: Prognostic variables in newly diagnosed children and adolescents with acute myeloid leukemia: Children's Cancer Group Study 213. Leukemia 16 (4): 601-7, 2002.
-
Hasle H, Alonzo TA, Auvrignon A, et al.: Monosomy 7 and deletion 7q in children and adolescents with acute myeloid leukemia: an international retrospective study. Blood 109 (11): 4641-7, 2007.
-
Swansbury GJ, Lawler SD, Alimena G, et al.: Long-term survival in acute myelogenous leukemia: a second follow-up of the Fourth International Workshop on Chromosomes in Leukemia. Cancer Genet Cytogenet 73 (1): 1-7, 1994.
-
Blink M, Zimmermann M, von Neuhoff C, et al.: Normal karyotype is a poor prognostic factor in myeloid leukemia of Down syndrome: a retrospective, international study. Haematologica 99 (2): 299-307, 2014.
-
Lugthart S, Gröschel S, Beverloo HB, et al.: Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol 28 (24): 3890-8, 2010.
-
Balgobind BV, Lugthart S, Hollink IH, et al.: EVI1 overexpression in distinct subtypes of pediatric acute myeloid leukemia. Leukemia 24 (5): 942-9, 2010.
-
Schnittger S, Schoch C, Dugas M, et al.: Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 100 (1): 59-66, 2002.
-
Thiede C, Steudel C, Mohr B, et al.: Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood 99 (12): 4326-35, 2002.
-
Whitman SP, Archer KJ, Feng L, et al.: Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a cancer and leukemia group B study. Cancer Res 61 (19): 7233-9, 2001.
-
Iwai T, Yokota S, Nakao M, et al.: Internal tandem duplication of the FLT3 gene and clinical evaluation in childhood acute myeloid leukemia. The Children's Cancer and Leukemia Study Group, Japan. Leukemia 13 (1): 38-43, 1999.
-
Arrigoni P, Beretta C, Silvestri D, et al.: FLT3 internal tandem duplication in childhood acute myeloid leukaemia: association with hyperleucocytosis in acute promyelocytic leukaemia. Br J Haematol 120 (1): 89-92, 2003.
-
Meshinchi S, Stirewalt DL, Alonzo TA, et al.: Activating mutations of RTK/ras signal transduction pathway in pediatric acute myeloid leukemia. Blood 102 (4): 1474-9, 2003.
-
Zwaan CM, Meshinchi S, Radich JP, et al.: FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: prognostic significance and relation to cellular drug resistance. Blood 102 (7): 2387-94, 2003.
-
Meshinchi S, Alonzo TA, Stirewalt DL, et al.: Clinical implications of FLT3 mutations in pediatric AML. Blood 108 (12): 3654-61, 2006.
-
Chang P, Kang M, Xiao A, et al.: FLT3 mutation incidence and timing of origin in a population case series of pediatric leukemia. BMC Cancer 10: 513, 2010.
-
Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, et al.: NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 118 (13): 3645-56, 2011.
-
Ostronoff F, Othus M, Gerbing RB, et al.: NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood 124 (15): 2400-7, 2014.
-
Shih LY, Kuo MC, Liang DC, et al.: Internal tandem duplication and Asp835 mutations of the FMS-like tyrosine kinase 3 (FLT3) gene in acute promyelocytic leukemia. Cancer 98 (6): 1206-16, 2003.
-
Noguera NI, Breccia M, Divona M, et al.: Alterations of the FLT3 gene in acute promyelocytic leukemia: association with diagnostic characteristics and analysis of clinical outcome in patients treated with the Italian AIDA protocol. Leukemia 16 (11): 2185-9, 2002.
-
Gale RE, Hills R, Pizzey AR, et al.: Relationship between FLT3 mutation status, biologic characteristics, and response to targeted therapy in acute promyelocytic leukemia. Blood 106 (12): 3768-76, 2005.
-
Abu-Duhier FM, Goodeve AC, Wilson GA, et al.: Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br J Haematol 113 (4): 983-8, 2001.
-
Tallman MS, Kim HT, Montesinos P, et al.: Does microgranular variant morphology of acute promyelocytic leukemia independently predict a less favorable outcome compared with classical M3 APL? A joint study of the North American Intergroup and the PETHEMA Group. Blood 116 (25): 5650-9, 2010.
-
Sung L, Aplenc R, Alonzo TA, et al.: Predictors and short-term outcomes of hyperleukocytosis in children with acute myeloid leukemia: a report from the Children's Oncology Group. Haematologica 97 (11): 1770-3, 2012.
-
Callens C, Chevret S, Cayuela JM, et al.: Prognostic implication of FLT3 and Ras gene mutations in patients with acute promyelocytic leukemia (APL): a retrospective study from the European APL Group. Leukemia 19 (7): 1153-60, 2005.
-
Schnittger S, Bacher U, Haferlach C, et al.: Clinical impact of FLT3 mutation load in acute promyelocytic leukemia with t(15;17)/PML-RARA. Haematologica 96 (12): 1799-807, 2011.
-
Pui CH, Relling MV, Rivera GK, et al.: Epipodophyllotoxin-related acute myeloid leukemia: a study of 35 cases. Leukemia 9 (12): 1990-6, 1995.
-
Inaba H, Zhou Y, Abla O, et al.: Heterogeneous cytogenetic subgroups and outcomes in childhood acute megakaryoblastic leukemia: a retrospective international study. Blood 126 (13): 1575-84, 2015.
-
Balgobind BV, Raimondi SC, Harbott J, et al.: Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood 114 (12): 2489-96, 2009.
-
Swansbury GJ, Slater R, Bain BJ, et al.: Hematological malignancies with t(9;11)(p21-22;q23)--a laboratory and clinical study of 125 cases. European 11q23 Workshop participants. Leukemia 12 (5): 792-800, 1998.
-
Rubnitz JE, Raimondi SC, Tong X, et al.: Favorable impact of the t(9;11) in childhood acute myeloid leukemia. J Clin Oncol 20 (9): 2302-9, 2002.
-
Mrózek K, Heinonen K, Lawrence D, et al.: Adult patients with de novo acute myeloid leukemia and t(9; 11)(p22; q23) have a superior outcome to patients with other translocations involving band 11q23: a Cancer and Leukemia Group B study. Blood 90 (11): 4532-8, 1997.
-
Martinez-Climent JA, Espinosa R 3rd, Thirman MJ, et al.: Abnormalities of chromosome band 11q23 and the MLL gene in pediatric myelomonocytic and monoblastic leukemias. Identification of the t(9;11) as an indicator of long survival. J Pediatr Hematol Oncol 17 (4): 277-83, 1995.
-
Casillas JN, Woods WG, Hunger SP, et al.: Prognostic implications of t(10;11) translocations in childhood acute myelogenous leukemia: a report from the Children's Cancer Group. J Pediatr Hematol Oncol 25 (8): 594-600, 2003.
-
Morerio C, Rosanda C, Rapella A, et al.: Is t(10;11)(p11.2;q23) involving MLL and ABI-1 genes associated with congenital acute monocytic leukemia? Cancer Genet Cytogenet 139 (1): 57-9, 2002.
-
Taki T, Shibuya N, Taniwaki M, et al.: ABI-1, a human homolog to mouse Abl-interactor 1, fuses the MLL gene in acute myeloid leukemia with t(10;11)(p11.2;q23). Blood 92 (4): 1125-30, 1998.
-
Coenen EA, Raimondi SC, Harbott J, et al.: Prognostic significance of additional cytogenetic aberrations in 733 de novo pediatric 11q23/MLL-rearranged AML patients: results of an international study. Blood 117 (26): 7102-11, 2011.
-
Ageberg M, Drott K, Olofsson T, et al.: Identification of a novel and myeloid specific role of the leukemia-associated fusion protein DEK-NUP214 leading to increased protein synthesis. Genes Chromosomes Cancer 47 (4): 276-87, 2008.
-
Shiba N, Ichikawa H, Taki T, et al.: NUP98-NSD1 gene fusion and its related gene expression signature are strongly associated with a poor prognosis in pediatric acute myeloid leukemia. Genes Chromosomes Cancer 52 (7): 683-93, 2013.
-
Slovak ML, Gundacker H, Bloomfield CD, et al.: A retrospective study of 69 patients with t(6;9)(p23;q34) AML emphasizes the need for a prospective, multicenter initiative for rare 'poor prognosis' myeloid malignancies. Leukemia 20 (7): 1295-7, 2006.
-
Alsabeh R, Brynes RK, Slovak ML, et al.: Acute myeloid leukemia with t(6;9) (p23;q34): association with myelodysplasia, basophilia, and initial CD34 negative immunophenotype. Am J Clin Pathol 107 (4): 430-7, 1997.
-
Sandahl JD, Coenen EA, Forestier E, et al.: t(6;9)(p22;q34)/DEK-NUP214-rearranged pediatric myeloid leukemia: an international study of 62 patients. Haematologica 99 (5): 865-72, 2014.
-
Tarlock K, Alonzo TA, Moraleda PP, et al.: Acute myeloid leukaemia (AML) with t(6;9)(p23;q34) is associated with poor outcome in childhood AML regardless of FLT3-ITD status: a report from the Children's Oncology Group. Br J Haematol 166 (2): 254-9, 2014.
-
Carroll A, Civin C, Schneider N, et al.: The t(1;22) (p13;q13) is nonrandom and restricted to infants with acute megakaryoblastic leukemia: a Pediatric Oncology Group Study. Blood 78 (3): 748-52, 1991.
-
Lion T, Haas OA: Acute megakaryocytic leukemia with the t(1;22)(p13;q13). Leuk Lymphoma 11 (1-2): 15-20, 1993.
-
Duchayne E, Fenneteau O, Pages MP, et al.: Acute megakaryoblastic leukaemia: a national clinical and biological study of 53 adult and childhood cases by the Groupe Français d'Hématologie Cellulaire (GFHC). Leuk Lymphoma 44 (1): 49-58, 2003.
-
de Rooij JD, Masetti R, van den Heuvel-Eibrink MM, et al.: Recurrent abnormalities can be used for risk group stratification in pediatric AMKL: a retrospective intergroup study. Blood 127 (26): 3424-30, 2016.
-
de Rooij JD, Hollink IH, Arentsen-Peters ST, et al.: NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia 27 (12): 2280-8, 2013.
-
Bernstein J, Dastugue N, Haas OA, et al.: Nineteen cases of the t(1;22)(p13;q13) acute megakaryblastic leukaemia of infants/children and a review of 39 cases: report from a t(1;22) study group. Leukemia 14 (1): 216-8, 2000.
-
Ma Z, Morris SW, Valentine V, et al.: Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet 28 (3): 220-1, 2001.
-
Mercher T, Coniat MB, Monni R, et al.: Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci U S A 98 (10): 5776-9, 2001.
-
Schweitzer J, Zimmermann M, Rasche M, et al.: Improved outcome of pediatric patients with acute megakaryoblastic leukemia in the AML-BFM 04 trial. Ann Hematol 94 (8): 1327-36, 2015.
-
Coenen EA, Zwaan CM, Reinhardt D, et al.: Pediatric acute myeloid leukemia with t(8;16)(p11;p13), a distinct clinical and biological entity: a collaborative study by the International-Berlin-Frankfurt-Munster AML-study group. Blood 122 (15): 2704-13, 2013.
-
Wong KF, Yuen HL, Siu LL, et al.: t(8;16)(p11;p13) predisposes to a transient but potentially recurring neonatal leukemia. Hum Pathol 39 (11): 1702-7, 2008.
-
Wu X, Sulavik D, Roulston D, et al.: Spontaneous remission of congenital acute myeloid leukemia with t(8;16)(p11;13). Pediatr Blood Cancer 56 (2): 331-2, 2011.
-
Terui K, Sato T, Sasaki S, et al.: Two novel variants of MOZ-CBP fusion transcripts in spontaneously remitted infant leukemia with t(1;16;8)(p13;p13;p11), a new variant of t(8;16)(p11;p13). Haematologica 93 (10): 1591-3, 2008.
-
Sainati L, Bolcato S, Cocito MG, et al.: Transient acute monoblastic leukemia with reciprocal (8;16)(p11;p13) translocation. Pediatr Hematol Oncol 13 (2): 151-7, 1996 Mar-Apr.
-
Weintraub M, Kaplinsky C, Amariglio N, et al.: Spontaneous regression of congenital leukaemia with an 8;16 translocation. Br J Haematol 111 (2): 641-3, 2000.
-
Classen CF, Behnisch W, Reinhardt D, et al.: Spontaneous complete and sustained remission of a rearrangement CBP (16p13)-positive disseminated congenital myelosarcoma. Ann Hematol 84 (4): 274-5, 2005.
-
Beverloo HB, Panagopoulos I, Isaksson M, et al.: Fusion of the homeobox gene HLXB9 and the ETV6 gene in infant acute myeloid leukemias with the t(7;12)(q36;p13). Cancer Res 61 (14): 5374-7, 2001.
-
Slater RM, von Drunen E, Kroes WG, et al.: t(7;12)(q36;p13) and t(7;12)(q32;p13)--translocations involving ETV6 in children 18 months of age or younger with myeloid disorders. Leukemia 15 (6): 915-20, 2001.
-
von Bergh AR, van Drunen E, van Wering ER, et al.: High incidence of t(7;12)(q36;p13) in infant AML but not in infant ALL, with a dismal outcome and ectopic expression of HLXB9. Genes Chromosomes Cancer 45 (8): 731-9, 2006.
-
Tosi S, Harbott J, Teigler-Schlegel A, et al.: t(7;12)(q36;p13), a new recurrent translocation involving ETV6 in infant leukemia. Genes Chromosomes Cancer 29 (4): 325-32, 2000.
-
Park J, Kim M, Lim J, et al.: Three-way complex translocations in infant acute myeloid leukemia with t(7;12)(q36;p13): the incidence and correlation of a HLXB9 overexpression. Cancer Genet Cytogenet 191 (2): 102-5, 2009.
-
Takeda A, Yaseen NR: Nucleoporins and nucleocytoplasmic transport in hematologic malignancies. Semin Cancer Biol 27: 3-10, 2014.
-
Brown J, Jawad M, Twigg SR, et al.: A cryptic t(5;11)(q35;p15.5) in 2 children with acute myeloid leukemia with apparently normal karyotypes, identified by a multiplex fluorescence in situ hybridization telomere assay. Blood 99 (7): 2526-31, 2002.
-
Panarello C, Rosanda C, Morerio C: Cryptic translocation t(5;11)(q35;p15.5) with involvement of the NSD1 and NUP98 genes without 5q deletion in childhood acute myeloid leukemia. Genes Chromosomes Cancer 35 (3): 277-81, 2002.
-
Cerveira N, Correia C, Dória S, et al.: Frequency of NUP98-NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia 17 (11): 2244-7, 2003.
-
Jaju RJ, Fidler C, Haas OA, et al.: A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 98 (4): 1264-7, 2001.
-
Gruber TA, Larson Gedman A, Zhang J, et al.: An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 22 (5): 683-97, 2012.
-
Thiollier C, Lopez CK, Gerby B, et al.: Characterization of novel genomic alterations and therapeutic approaches using acute megakaryoblastic leukemia xenograft models. J Exp Med 209 (11): 2017-31, 2012.
-
Masetti R, Pigazzi M, Togni M, et al.: CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood 121 (17): 3469-72, 2013.
-
Masetti R, Rondelli R, Fagioli F, et al.: Infants with acute myeloid leukemia treated according to the Associazione Italiana di Ematologia e Oncologia Pediatrica 2002/01 protocol have an outcome comparable to that of older children. Haematologica 99 (8): e127-9, 2014.
-
Radich JP, Kopecky KJ, Willman CL, et al.: N-ras mutations in adult de novo acute myelogenous leukemia: prevalence and clinical significance. Blood 76 (4): 801-7, 1990.
-
Farr C, Gill R, Katz F, et al.: Analysis of ras gene mutations in childhood myeloid leukaemia. Br J Haematol 77 (3): 323-7, 1991.
-
Berman JN, Gerbing RB, Alonzo TA, et al.: Prevalence and clinical implications of NRAS mutations in childhood AML: a report from the Children's Oncology Group. Leukemia 25 (6): 1039-42, 2011.
-
Schnittger S, Kohl TM, Haferlach T, et al.: KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood 107 (5): 1791-9, 2006.
-
Tokumasu M, Murata C, Shimada A, et al.: Adverse prognostic impact of KIT mutations in childhood CBF-AML: the results of the Japanese Pediatric Leukemia/Lymphoma Study Group AML-05 trial. Leukemia 29 (12): 2438-41, 2015.
-
Cairoli R, Beghini A, Grillo G, et al.: Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood 107 (9): 3463-8, 2006.
-
Paschka P, Marcucci G, Ruppert AS, et al.: Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol 24 (24): 3904-11, 2006.
-
Shimada A, Taki T, Tabuchi K, et al.: KIT mutations, and not FLT3 internal tandem duplication, are strongly associated with a poor prognosis in pediatric acute myeloid leukemia with t(8;21): a study of the Japanese Childhood AML Cooperative Study Group. Blood 107 (5): 1806-9, 2006.
-
Shih LY, Liang DC, Huang CF, et al.: Cooperating mutations of receptor tyrosine kinases and Ras genes in childhood core-binding factor acute myeloid leukemia and a comparative analysis on paired diagnosis and relapse samples. Leukemia 22 (2): 303-7, 2008.
-
Goemans BF, Zwaan CM, Miller M, et al.: Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia 19 (9): 1536-42, 2005.
-
Boissel N, Leroy H, Brethon B, et al.: Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML). Leukemia 20 (6): 965-70, 2006.
-
Pollard JA, Alonzo TA, Gerbing RB, et al.: Prevalence and prognostic significance of KIT mutations in pediatric patients with core binding factor AML enrolled on serial pediatric cooperative trials for de novo AML. Blood 115 (12): 2372-9, 2010.
-
Groet J, McElwaine S, Spinelli M, et al.: Acquired mutations in GATA1 in neonates with Down's syndrome with transient myeloid disorder. Lancet 361 (9369): 1617-20, 2003.
-
Hitzler JK, Cheung J, Li Y, et al.: GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood 101 (11): 4301-4, 2003.
-
Rainis L, Bercovich D, Strehl S, et al.: Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood 102 (3): 981-6, 2003.
-
Wechsler J, Greene M, McDevitt MA, et al.: Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet 32 (1): 148-52, 2002.
-
Gurbuxani S, Vyas P, Crispino JD: Recent insights into the mechanisms of myeloid leukemogenesis in Down syndrome. Blood 103 (2): 399-406, 2004.
-
Ge Y, Stout ML, Tatman DA, et al.: GATA1, cytidine deaminase, and the high cure rate of Down syndrome children with acute megakaryocytic leukemia. J Natl Cancer Inst 97 (3): 226-31, 2005.
-
Paschka P, Marcucci G, Ruppert AS, et al.: Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol 26 (28): 4595-602, 2008.
-
Virappane P, Gale R, Hills R, et al.: Mutation of the Wilms' tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukemia: the United Kingdom Medical Research Council Adult Leukaemia Working Party. J Clin Oncol 26 (33): 5429-35, 2008.
-
Gaidzik VI, Schlenk RF, Moschny S, et al.: Prognostic impact of WT1 mutations in cytogenetically normal acute myeloid leukemia: a study of the German-Austrian AML Study Group. Blood 113 (19): 4505-11, 2009.
-
Renneville A, Boissel N, Zurawski V, et al.: Wilms tumor 1 gene mutations are associated with a higher risk of recurrence in young adults with acute myeloid leukemia: a study from the Acute Leukemia French Association. Cancer 115 (16): 3719-27, 2009.
-
Ho PA, Zeng R, Alonzo TA, et al.: Prevalence and prognostic implications of WT1 mutations in pediatric acute myeloid leukemia (AML): a report from the Children's Oncology Group. Blood 116 (5): 702-10, 2010.
-
Hollink IH, van den Heuvel-Eibrink MM, Zimmermann M, et al.: Clinical relevance of Wilms tumor 1 gene mutations in childhood acute myeloid leukemia. Blood 113 (23): 5951-60, 2009.
-
Ley TJ, Ding L, Walter MJ, et al.: DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363 (25): 2424-33, 2010.
-
Yan XJ, Xu J, Gu ZH, et al.: Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 43 (4): 309-15, 2011.
-
Thol F, Damm F, Lüdeking A, et al.: Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol 29 (21): 2889-96, 2011.
-
Green CL, Evans CM, Hills RK, et al.: The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood 116 (15): 2779-82, 2010.
-
Paschka P, Schlenk RF, Gaidzik VI, et al.: IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 28 (22): 3636-43, 2010.
-
Abbas S, Lugthart S, Kavelaars FG, et al.: Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood 116 (12): 2122-6, 2010.
-
Marcucci G, Maharry K, Wu YZ, et al.: IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 28 (14): 2348-55, 2010.
-
Wagner K, Damm F, Göhring G, et al.: Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol 28 (14): 2356-64, 2010.
-
Figueroa ME, Abdel-Wahab O, Lu C, et al.: Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18 (6): 553-67, 2010.
-
Ward PS, Patel J, Wise DR, et al.: The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17 (3): 225-34, 2010.
-
Dang L, White DW, Gross S, et al.: Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462 (7274): 739-44, 2009.
-
Damm F, Thol F, Hollink I, et al.: Prevalence and prognostic value of IDH1 and IDH2 mutations in childhood AML: a study of the AML-BFM and DCOG study groups. Leukemia 25 (11): 1704-10, 2011.
-
Oki K, Takita J, Hiwatari M, et al.: IDH1 and IDH2 mutations are rare in pediatric myeloid malignancies. Leukemia 25 (2): 382-4, 2011.
-
Pigazzi M, Ferrari G, Masetti R, et al.: Low prevalence of IDH1 gene mutation in childhood AML in Italy. Leukemia 25 (1): 173-4, 2011.
-
Ho PA, Alonzo TA, Kopecky KJ, et al.: Molecular alterations of the IDH1 gene in AML: a Children's Oncology Group and Southwest Oncology Group study. Leukemia 24 (5): 909-13, 2010.
-
Andersson AK, Miller DW, Lynch JA, et al.: IDH1 and IDH2 mutations in pediatric acute leukemia. Leukemia 25 (10): 1570-7, 2011.
-
Maxson JE, Ries RE, Wang YC, et al.: CSF3R mutations have a high degree of overlap with CEBPA mutations in pediatric AML. Blood 127 (24): 3094-8, 2016.
-
Germeshausen M, Kratz CP, Ballmaier M, et al.: RAS and CSF3R mutations in severe congenital neutropenia. Blood 114 (16): 3504-5, 2009.
-
Skokowa J, Steinemann D, Katsman-Kuipers JE, et al.: Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood 123 (14): 2229-37, 2014.
-
Joyce CE, Novina CD: miR-155 in acute myeloid leukemia: not merely a prognostic marker? J Clin Oncol 31 (17): 2219-21, 2013.
-
Marcucci G, Maharry KS, Metzeler KH, et al.: Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol 31 (17): 2086-93, 2013.
-
Ramamurthy R, Hughes M, Morris V, et al.: miR-155 expression and correlation with clinical outcome in pediatric AML: A report from Children's Oncology Group. Pediatr Blood Cancer 63 (12): 2096-2103, 2016.
-
Caye A, Strullu M, Guidez F, et al.: Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat Genet 47 (11): 1334-40, 2015.
-
Stieglitz E, Taylor-Weiner AN, Chang TY, et al.: The genomic landscape of juvenile myelomonocytic leukemia. Nat Genet 47 (11): 1326-33, 2015.
-
Sakaguchi H, Okuno Y, Muramatsu H, et al.: Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet 45 (8): 937-41, 2013.
-
Hernández-Porras I, Fabbiano S, Schuhmacher AJ, et al.: K-RasV14I recapitulates Noonan syndrome in mice. Proc Natl Acad Sci U S A 111 (46): 16395-400, 2014.
-
Chang T, Krisman K, Theobald EH, et al.: Sustained MEK inhibition abrogates myeloproliferative disease in Nf1 mutant mice. J Clin Invest 123 (1): 335-9, 2013.
Non-Hodgkin LymphomaMature B-cell Lymphoma The mature B-cell lymphomas include Burkitt and Burkitt-like lymphoma, diffuse large B-cell lymphoma, and primary mediastinal B-cell lymphoma. Burkitt and Burkitt-like lymphoma The malignant cells show a mature B-cell phenotype and are negative for the enzyme terminal deoxynucleotidyl transferase. These malignant cells usually express surface immunoglobulin, most bearing a clonal surface immunoglobulin M with either kappa or lambda light chains. A variety of additional B-cell markers (e.g., CD19, CD20, CD22) are usually present, and most childhood Burkitt and Burkitt-like lymphomas/leukemias express CALLA (CD10).[1] Burkitt lymphoma/leukemia expresses a characteristic chromosomal translocation, usually t(8;14) and more rarely t(8;22) or t(2;8). Each of these translocations juxtaposes the c-myc oncogene and immunoglobulin locus regulatory elements, resulting in the inappropriate expression of c-myc, a gene involved in cellular proliferation.[2,3,4] The presence of one of the variant translocations t(2;8) or t(8;22) does not appear to affect response or outcome.[5] The distinction between Burkitt and Burkitt-like lymphoma/leukemia is controversial. Burkitt lymphoma/leukemia consists of uniform, small, noncleaved cells, whereas the diagnosis of Burkitt-like lymphoma/leukemia is highly disputed among pathologists because of features that are consistent with diffuse large B-cell lymphoma.[6] Cytogenetic evidence of c-myc rearrangement is the gold standard for diagnosis of Burkitt lymphoma/leukemia. For cases in which cytogenetic analysis is not available, the World Health Organization (WHO) has recommended that the Burkitt-like diagnosis be reserved for lymphoma resembling Burkitt lymphoma/leukemia or with more pleomorphism, large cells, and a proliferation fraction (i.e., MIB-1 or Ki-67 immunostaining) of 99% or greater.[1] BCL2 staining by immunohistochemistry is variable. The absence of a translocation involving the BCL2 gene does not preclude the diagnosis of Burkitt lymphoma/leukemia and has no clinical implications.[7] Studies have demonstrated that the vast majority of Burkitt-like or atypical Burkitt lymphoma/leukemia has a gene expression signature similar to Burkitt lymphoma/leukemia.[8,9] Additionally, as many as 30% of pediatric diffuse large B-cell lymphoma cases will have a gene signature similar to Burkitt lymphoma/leukemia.[8,10] (Refer to the PDQ summary on Childhood Non-Hodgkin Lymphoma Treatment for information about the treatment of childhood non-Hodgkin lymphoma.) Diffuse large B-cell lymphoma The World Health Organization (WHO) classification system does not recommend subclassification of diffuse large B-cell lymphoma on the basis of morphologic variants (e.g., immunoblastic, centroblastic).[11] Diffuse large B-cell lymphoma in children and adolescents differs biologically from diffuse large B-cell lymphoma in adults in the following ways: - The vast majority of pediatric diffuse large B-cell lymphoma cases have a germinal center B-cell phenotype, as assessed by immunohistochemical analysis of selected proteins found in normal germinal center B cells, such as the BCL6 gene product and CD10.[5,12,13] The age at which the favorable germinal center subtype changes to the less favorable nongerminal center subtype was shown to be a continuous variable.[14]
- Pediatric diffuse large B-cell lymphoma rarely demonstrates the t(14;18) translocation involving the immunoglobulin heavy-chain gene and the BCL2 gene that is seen adults.[12]
- As many as 30% of patients younger than 14 years with diffuse large B-cell lymphoma will have a gene signature similar to Burkitt lymphoma/leukemia.[8,10]
- In contrast to adult diffuse large B-cell lymphoma, pediatric cases show a high frequency of abnormalities at the MYC locus (chromosome 8q24), with approximately one-third of pediatric cases showing MYC rearrangement and with approximately one-half of the nonrearranged cases showing MYC gain or amplification.[10,15]
- A subset of pediatric diffuse large B-cell lymphoma cases was found to have a translocation that juxtaposes the IRF4 oncogene next to one of the immunoglobulin loci. Diffuse large B-cell lymphoma cases with an IRF4 translocation were significantly more frequent in children than in adults (15% vs. 2%), were germinal center-derived B-cell lymphomas, and were associated with favorable prognosis compared with diffuse large B-cell lymphoma cases lacking this abnormality.[16]
(Refer to the PDQ summary on Childhood Non-Hodgkin Lymphoma Treatment for information about the treatment of childhood non-Hodgkin lymphoma.) Primary mediastinal B-cell lymphoma Primary mediastinal B-cell lymphoma was previously considered a subtype of diffuse large B-cell lymphoma, but is now a separate entity in the most recent World Health Organization (WHO) classification.[17] These tumors arise in the mediastinum from thymic B-cells and show a diffuse large cell proliferation with sclerosis that compartmentalizes neoplastic cells. Primary mediastinal B-cell lymphoma can be very difficult to distinguish morphologically from the following types of lymphoma: - Diffuse large B-cell lymphoma: Cell surface markers are similar to the ones seen in diffuse large B-cell lymphoma, such as CD19, CD20, CD22, CD79a, and PAX-5. Primary mediastinal B-cell lymphoma often lacks cell surface immunoglobulin expression but may display cytoplasmic immunoglobulins. CD30 expression is commonly present.[17]
- Hodgkin lymphoma: Primary mediastinal B-cell lymphoma may be difficult to clinically and morphologically distinguish from Hodgkin lymphoma, especially with small mediastinal biopsies because of extensive sclerosis and necrosis.
Primary mediastinal B-cell lymphoma is associated with distinctive chromosomal aberrations (gains in chromosomes 9p and 2p in regions that involve JAK2 and c-rel, respectively) [18,19] and commonly shows inactivation of SOCS1 by either mutation or gene deletion.[20,21] Primary mediastinal B-cell lymphoma has a distinctly different gene expression profile from diffuse large B-cell lymphoma, but its gene expression profile has features similar to those seen in Hodgkin lymphoma.[22,23] (Refer to the PDQ summary on Childhood Non-Hodgkin Lymphoma Treatment for information about the treatment of childhood non-Hodgkin lymphoma.) Lymphoblastic Lymphoma Lymphoblastic lymphomas are usually positive for terminal deoxynucleotidyl transferase, with more than 75% having a T-cell immunophenotype and the remainder having a precursor B-cell phenotype.[2,24] As opposed to pediatric acute lymphoblastic leukemia, chromosomal abnormalities and the molecular biology of pediatric lymphoblastic lymphoma are not well characterized. The Berlin-Frankfurt-Münster group reported that loss of heterozygosity at chromosome 6q was observed in 12% of patients and NOTCH1 mutations were seen in 60% of patients, but NOTCH1 mutations are rarely seen in patients with loss of heterozygosity in 6q16.[25,26] (Refer to the PDQ summary on Childhood Non-Hodgkin Lymphoma Treatment for information about the treatment of childhood non-Hodgkin lymphoma.) Anaplastic Large Cell Lymphoma While the predominant immunophenotype of anaplastic large cell lymphoma is mature T-cell, null-cell disease (i.e., no T-cell, B-cell, or natural killer-cell surface antigen expression) does occur. The World Health Organization (WHO) classifies anaplastic large cell lymphoma as a subtype of peripheral T-cell lymphoma.[27] All anaplastic large cell lymphoma cases are CD30-positive. More than 90% of pediatric anaplastic large cell lymphoma cases have a chromosomal rearrangement involving the ALK gene. About 85% of these chromosomal rearrangements will be t(2;5)(p23;q35), leading to the expression of the fusion protein NPM-ALK; the other 15% of cases are composed of variant ALK translocations.[28] Anti-ALK immunohistochemical staining pattern is quite specific for the type of ALK translocation. Cytoplasm and nuclear ALK staining is associated with NPM-ALK fusion protein, whereas cytoplasmic staining only of ALK is associated with the variant ALK translocations.[28] In adults, ALK-positive anaplastic large cell lymphoma is viewed differently from other peripheral T-cell lymphomas because prognosis tends to be superior.[29] Also, adult ALK-negative anaplastic large cell lymphoma patients have an inferior outcome compared with patients who have ALK-positive disease.[30] In children, however, this difference in outcome between ALK-positive and ALK-negative disease has not been demonstrated. In addition, no correlation has been found between outcome and the specific ALK-translocation type.[31,32,33] In a European series of 375 children and adolescents with systemic ALK-positive anaplastic large cell lymphoma, the presence of a small cell or lymphohistiocytic component was observed in 32% of patients and was significantly associated with a high risk of failure in the multivariate analysis, controlling for clinical characteristics (hazard ratio, 2.0; P = .002).[32] The prognostic implication of the small cell variant of anaplastic large cell lymphoma was also shown in the COG-ANHL0131 (NCT00059839) study, despite a different chemotherapy backbone.[33] (Refer to the PDQ summary on Childhood Non-Hodgkin Lymphoma Treatment for information about the treatment of childhood non-Hodgkin lymphoma.) References:
-
Leoncini L, Raphael M, Stein H: Burkitt lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., eds.: WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: International Agency for Research on Cancer, 2008, pp 262-4.
-
Sandlund JT, Downing JR, Crist WM: Non-Hodgkin's lymphoma in childhood. N Engl J Med 334 (19): 1238-48, 1996.
-
Perkins SL, Lones MA, Davenport V, et al.: B-Cell non-Hodgkin's lymphoma in children and adolescents: surface antigen expression and clinical implications for future targeted bioimmune therapy: a children's cancer group report. Clin Adv Hematol Oncol 1 (5): 314-7, 2003.
-
Miles RR, Cairo MS, Satwani P, et al.: Immunophenotypic identification of possible therapeutic targets in paediatric non-Hodgkin lymphomas: a children's oncology group report. Br J Haematol 138 (4): 506-12, 2007.
-
Gualco G, Weiss LM, Harrington WJ Jr, et al.: Nodal diffuse large B-cell lymphomas in children and adolescents: immunohistochemical expression patterns and c-MYC translocation in relation to clinical outcome. Am J Surg Pathol 33 (12): 1815-22, 2009.
-
Kluin PM, Harris NL, Stein H: B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., eds.: WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: International Agency for Research on Cancer, 2008, pp 265-6.
-
Masqué-Soler N, Szczepanowski M, Kohler CW, et al.: Clinical and pathological features of Burkitt lymphoma showing expression of BCL2--an analysis including gene expression in formalin-fixed paraffin-embedded tissue. Br J Haematol 171 (4): 501-8, 2015.
-
Klapper W, Szczepanowski M, Burkhardt B, et al.: Molecular profiling of pediatric mature B-cell lymphoma treated in population-based prospective clinical trials. Blood 112 (4): 1374-81, 2008.
-
Dave SS, Fu K, Wright GW, et al.: Molecular diagnosis of Burkitt's lymphoma. N Engl J Med 354 (23): 2431-42, 2006.
-
Deffenbacher KE, Iqbal J, Sanger W, et al.: Molecular distinctions between pediatric and adult mature B-cell non-Hodgkin lymphomas identified through genomic profiling. Blood 119 (16): 3757-66, 2012.
-
Stein H, Warnke RA, Chan WC: Diffuse large B-cell lymphoma (DLBCL), NOS. In: Swerdlow SH, Campo E, Harris NL, et al., eds.: WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: International Agency for Research on Cancer, 2008, pp 233-7.
-
Oschlies I, Klapper W, Zimmermann M, et al.: Diffuse large B-cell lymphoma in pediatric patients belongs predominantly to the germinal-center type B-cell lymphomas: a clinicopathologic analysis of cases included in the German BFM (Berlin-Frankfurt-Munster) Multicenter Trial. Blood 107 (10): 4047-52, 2006.
-
Miles RR, Raphael M, McCarthy K, et al.: Pediatric diffuse large B-cell lymphoma demonstrates a high proliferation index, frequent c-Myc protein expression, and a high incidence of germinal center subtype: Report of the French-American-British (FAB) international study group. Pediatr Blood Cancer 51 (3): 369-74, 2008.
-
Klapper W, Kreuz M, Kohler CW, et al.: Patient age at diagnosis is associated with the molecular characteristics of diffuse large B-cell lymphoma. Blood 119 (8): 1882-7, 2012.
-
Poirel HA, Cairo MS, Heerema NA, et al.: Specific cytogenetic abnormalities are associated with a significantly inferior outcome in children and adolescents with mature B-cell non-Hodgkin's lymphoma: results of the FAB/LMB 96 international study. Leukemia 23 (2): 323-31, 2009.
-
Salaverria I, Philipp C, Oschlies I, et al.: Translocations activating IRF4 identify a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults. Blood 118 (1): 139-47, 2011.
-
Jaffe ES, Harris NL, Stein H, et al.: Introduction and overview of the classification of the lymphoid neoplasms. In: Swerdlow SH, Campo E, Harris NL, et al., eds.: WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: International Agency for Research on Cancer, 2008, pp 157-66.
-
Bea S, Zettl A, Wright G, et al.: Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood 106 (9): 3183-90, 2005.
-
Oschlies I, Burkhardt B, Salaverria I, et al.: Clinical, pathological and genetic features of primary mediastinal large B-cell lymphomas and mediastinal gray zone lymphomas in children. Haematologica 96 (2): 262-8, 2011.
-
Melzner I, Bucur AJ, Brüderlein S, et al.: Biallelic mutation of SOCS-1 impairs JAK2 degradation and sustains phospho-JAK2 action in the MedB-1 mediastinal lymphoma line. Blood 105 (6): 2535-42, 2005.
-
Mestre C, Rubio-Moscardo F, Rosenwald A, et al.: Homozygous deletion of SOCS1 in primary mediastinal B-cell lymphoma detected by CGH to BAC microarrays. Leukemia 19 (6): 1082-4, 2005.
-
Rosenwald A, Wright G, Leroy K, et al.: Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 198 (6): 851-62, 2003.
-
Savage KJ, Monti S, Kutok JL, et al.: The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood 102 (12): 3871-9, 2003.
-
Neth O, Seidemann K, Jansen P, et al.: Precursor B-cell lymphoblastic lymphoma in childhood and adolescence: clinical features, treatment, and results in trials NHL-BFM 86 and 90. Med Pediatr Oncol 35 (1): 20-7, 2000.
-
Bonn BR, Rohde M, Zimmermann M, et al.: Incidence and prognostic relevance of genetic variations in T-cell lymphoblastic lymphoma in childhood and adolescence. Blood 121 (16): 3153-60, 2013.
-
Burkhardt B, Moericke A, Klapper W, et al.: Pediatric precursor T lymphoblastic leukemia and lymphoblastic lymphoma: Differences in the common regions with loss of heterozygosity at chromosome 6q and their prognostic impact. Leuk Lymphoma 49 (3): 451-61, 2008.
-
Swerdlow SH, Campo E, Pileri SA, et al.: The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127 (20): 2375-90, 2016.
-
Duyster J, Bai RY, Morris SW: Translocations involving anaplastic lymphoma kinase (ALK). Oncogene 20 (40): 5623-37, 2001.
-
Savage KJ, Harris NL, Vose JM, et al.: ALK- anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-Cell Lymphoma Project. Blood 111 (12): 5496-504, 2008.
-
Vose J, Armitage J, Weisenburger D, et al.: International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol 26 (25): 4124-30, 2008.
-
Stein H, Foss HD, Dürkop H, et al.: CD30(+) anaplastic large cell lymphoma: a review of its histopathologic, genetic, and clinical features. Blood 96 (12): 3681-95, 2000.
-
Lamant L, McCarthy K, d'Amore E, et al.: Prognostic impact of morphologic and phenotypic features of childhood ALK-positive anaplastic large-cell lymphoma: results of the ALCL99 study. J Clin Oncol 29 (35): 4669-76, 2011.
-
Alexander S, Kraveka JM, Weitzman S, et al.: Advanced stage anaplastic large cell lymphoma in children and adolescents: results of ANHL0131, a randomized phase III trial of APO versus a modified regimen with vinblastine: a report from the children's oncology group. Pediatr Blood Cancer 61 (12): 2236-42, 2014.
Central Nervous System TumorsCentral nervous system (CNS) tumors include pilocytic astrocytomas and other astrocytic tumors, diffuse astrocytic tumors, brain stem gliomas, CNS atypical teratoid/rhabdoid tumors, medulloblastomas, nonmedulloblastoma embryonal tumors, and ependymomas. The terminology of the 2016 World Health Organization (WHO) Classification of Tumors of the Central Nervous System is used below. The 2016 WHO CNS classification incorporates genomic features in addition to histology, and it includes multiple changes from the previous 2007 WHO classification.[1] Of particular relevance for childhood brain cancers is the new entity diffuse midline glioma, H3 K27M-mutant, which includes diffuse intrinsic pontine glioma (DIPG) with the H3 K27M mutation and other high-grade gliomas of the midline with the H3 K27M mutation. Other examples of molecularly defined entities discussed below are RELA-fusion-positive ependymoma, WNT-activated and SHH-activated medulloblastoma, and embryonal tumor with multilayered rosettes, C19MC-altered. Pilocytic Astrocytomas and Other Astrocytic Tumors Genomic alterations involving activation of BRAF and the ERK/MAPK pathway are very common in sporadic cases of pilocytic astrocytoma, a type of low-grade glioma. BRAF activation in pilocytic astrocytoma occurs most commonly through a BRAF-KIAA1549 gene fusion, producing a fusion protein that lacks the BRAF regulatory domain.[2,3,4,5,6] This fusion is seen in most infratentorial and midline pilocytic astrocytomas, but is present at lower frequency in supratentorial (hemispheric) tumors.[2,3,7,8,9,10,11,12] Presence of the BRAF-KIAA1549 fusion predicted a better clinical outcome (progression-free survival [PFS] and overall survival [OS]) in one report that described children with incompletely resected low-grade gliomas.[11] However, other factors such as CDKN2A deletion, whole chromosome 7 gain, and tumor location may modify the impact of the BRAF mutation on outcome.[13]; [14][Level of evidence: 3iiiDiii] Progression to high-grade glioma is rare for pediatric low-grade glioma with the BRAF-KIAA1549 fusion.[15] BRAF activation through the BRAF-KIAA1549 fusion has also been described in other pediatric low-grade gliomas (e.g., pilomyxoid astrocytoma).[10,11] Other genomic alterations in pilocytic astrocytomas that can activate the ERK/MAPK pathway (e.g., alternative BRAF gene fusions, RAF1 rearrangements, RAS mutations, and BRAF V600E point mutations) are less commonly observed.[3,5,6,16]BRAF V600E point mutations are also observed in nonpilocytic pediatric low-grade gliomas, including ganglioglioma, desmoplastic infantile ganglioglioma, and approximately two-thirds of pleomorphic xanthoastrocytomas.[17,18,19] One retrospective study of 53 children with gangliogliomas demonstrated BRAF V600E staining in approximately 40% of tumors. Five-year recurrence-free survival was worse in the V600E-mutated tumors (about 60%) than in tumors that did not stain for V600E (about 80%).[20] Similarly, children with diencephalic low-grade astrocytomas with a BRAF V600E mutation had a 5-year PFS of 22%, compared with a 52% PFS in children who were BRAF wildtype.[21][Level of evidence: 3iiiDiii] The frequency of the BRAF V600E mutation was significantly higher in pediatric low-grade glioma that transformed to high-grade glioma (8 of 18 cases) than was the frequency of mutation in cases that did not transform (10 of 167 cases).[15] Angiocentric gliomas have been noted to largely harbor MYB-QKI fusions, a putative driver mutation for this relatively rare class of gliomas.[22] As with neurofibromatosis type 1 (NF1) deficiency in activating the ERK/MAPK pathway, activating BRAF genomic alterations are uncommon in pilocytic astrocytoma associated with NF1.[9] Activating mutations in FGFR1, PTPN11, and in NTRK2 fusion genes have also been identified in noncerebellar pilocytic astrocytomas.[23] In pediatric grade II diffuse astrocytomas, the most common alterations reported (up to 53% of tumors) are rearrangements in the MYB family of transcription factors.[24,25] Most children with tuberous sclerosis have a mutation in one of two tuberous sclerosis genes (TSC1/hamartin or TSC2/tuberin). Either of these mutations results in activation of the mammalian target of rapamycin (mTOR) complex 1. These children are at risk of developing subependymal giant cell astrocytomas, cortical tubers, and subependymal nodules. Because subependymal giant cell astrocytomas are driven by mTOR activation, mTOR inhibitors are active agents that can induce tumor regression in children with these tumors.[26] (Refer to the PDQ summary on Childhood Astrocytomas Treatment for information about the treatment of low-grade childhood astrocytomas.) Diffuse Astrocytic Tumors This category includes, among other diagnoses, diffuse astrocytomas (grade II) and pediatric high-grade gliomas (anaplastic astrocytoma [grade III], glioblastoma [grade IV], and diffuse midline glioma, H3 K27M-mutant (grade IV]). Diffuse astrocytomas For pediatric diffuse astrocytomas (grade II), rearrangements in the MYB family of transcription factors (MYB and MYBL1) are the most commonly reported genomic alteration.[24,25,27] Other alterations observed include FGFR1 alterations (primarily duplications involving the tyrosine kinase domain),[25,27]BRAF alterations, NF1 mutations, and RAS family mutations.[24,25]IDH1 mutations, which are the most common genomic alteration in adult diffuse astrocytomas, are uncommon in children with diffuse astrocytomas and, when present, are observed almost exclusively in older adolescents.[24,28] Anaplastic astrocytomas and glioblastomas Pediatric high-grade gliomas, especially glioblastoma multiforme, are biologically distinct from those arising in adults.[28,29,30,31] - Pediatric high-grade gliomas have PTEN and EGFR genomic alterations less frequently and PDGF/PDGFR genomic alterations and mutations in histone genes (primarily histone 3.3 [H3F3A] but also histone 3.1 [HIST1H3B]) more frequently than do adult tumors.
- Although it was believed that pediatric glioblastoma multiforme tumors were more closely related to adult secondary glioblastoma multiforme tumors in which there is stepwise transformation from lower-grade into higher-grade gliomas and in which most tumors have IDH1 and IDH2 mutations, the latter mutations are rarely observed in children younger than 15 years with high-grade gliomas.[32,33,34]IDH1 mutations are observed in older adolescents with high-grade gliomas.[28,34,35]
Pediatric glioblastoma multiforme tumors are separated into relatively distinct subgroups on the basis of epigenetic patterns (DNA methylation), with distinctive chromosome copy number gains/losses and gene mutations.[34,35] Two subgroups have identifiable recurrent mutations in H3F3A (the gene encoding histone 3.3), suggesting disrupted epigenetic regulatory mechanisms, with the most recognized subgroup having mutations at K27 (lysine 27) and the other group having mutations at G34 (glycine 34). The subgroups are the following: - H3F3A mutation at K27: The K27 cluster occurs predominately in mid-childhood (median age, approximately 10 years), is mainly midline (thalamus, brain stem, and spinal cord), and carries a very poor prognosis. These tumors also frequently have TP53 mutations. Thalamic high-grade gliomas in older adolescents and young adults also show a high rate of H3F3A K27 mutations.[36] The 2016 WHO classification groups these cancers into a single entity, diffuse midline glioma, H3 K27M-mutant.[1]
- H3F3A mutation at G34: The second H3F3A mutation tumor cluster, the G34 grouping, is found in somewhat older children and young adults (median age, 14-18 years), arises exclusively in the cerebral cortex, and carries a somewhat better prognosis.[34,35] The G34 clusters also have TP53 mutations and widespread hypomethylation across the whole genome. Patients with H3F3A mutations are at high risk of treatment failure, but the prognosis is not as poor as it is for patients with K27M mutations.[35]
The H3F3A K27 and G34 mutations appear to be unique to high-grade gliomas and have not been observed in other pediatric brain tumors.[37] Both mutations induce distinctive DNA methylation patterns compared with the patterns observed in IDH-mutated tumors, which occur in young adults.[32,33,34,37,38] Pediatric glioblastoma multiforme patients whose tumors have IDH1 mutations are almost exclusively older adolescents (median age in a pediatric population, 16 years) with hemispheric tumors. IDH1-mutated cases often show TP53 mutations, MGMT promoter methylation, and a glioma-CpG island methylator phenotype (G-CIMP).[34,35] Pediatric patients with IDH1 mutations show a more favorable prognosis than do other pediatric glioblastoma multiforme patients. A fourth group of pediatric glioblastoma multiforme patients identified by DNA methylation analysis are those lacking both histone mutations and IDH1 mutations. This is a heterogeneous group with higher rates of gene amplifications than other pediatric glioblastoma multiforme subtypes. The most commonly amplified genes are PDGFRA, EGFR, CCND/CDK, and MYC/MYCN.[34,35] DNA methylation analysis of tumor tissue may identify pediatric tumors with a histologic diagnosis of glioblastoma multiforme, but with the molecular characteristics of other pediatric gliomas. For example, one study found that approximately 14% of patients with a diagnosis of glioblastoma multiforme had molecular characteristics that are associated with pleomorphic xanthoastrocytomas (e.g., high rates of BRAF V600E mutations).[35] Infants and young children with a glioblastoma multiforme diagnosis appear to have tumors with distinctive molecular characteristics when compared with tumors of older children. One report that applied DNA methylation analysis to glioblastoma multiforme tumors observed a group of patients (representing approximately 7% of pediatric patients with a histologic diagnosis of glioblastoma multiforme) whose tumors had molecular characteristics consistent with low-grade gliomas. The median age for this group of patients was 1 year, and they showed a favorable prognosis (3-year overall survival, approximately 90%).[35] A second report investigated gene copy number gains and losses and mutation status of selected genes for glioblastoma multiforme tumors from children younger than 36 months.[39] Molecular alterations observed at appreciable rates in older children (e.g., K27M, CDKN2A loss, PDGFRA amplification, and TERT promoter mutations) were rare in the tumors of these young children, and novel abnormalities (e.g., loss of SNORD on chromosome 14q32) were observed in some cases. Childhood secondary high-grade glioma (high-grade glioma that is preceded by a low-grade glioma) is uncommon (2.9% in a study of 886 patients). No pediatric low-grade gliomas with the BRAF-KIAA1549 fusion transformed to a high-grade glioma, whereas low-grade gliomas with the BRAF V600E mutations were associated with increased risk of transformation. Seven of 18 patients (approximately 40%) with secondary high-grade glioma had BRAF V600E mutations, with CDKN2A alterations present in 8 of 14 cases (57%).[15] (Refer to the PDQ summary on Childhood Astrocytomas Treatment for information about the treatment of high-grade childhood astrocytomas.) Diffuse Midline Glioma,H3 K27M-Mutant (Including Diffuse Intrinsic Pontine Gliomas [DIPGs]) The diffuse midline glioma, H3 K27M-mutant, category includes tumors previously classified as DIPG; most of the data is derived from experience with DIPG. This category also includes gliomas with the H3 K27M mutation arising in midline structures such as the thalamus. The genomic characteristics of DIPGs appear to differ from those of most other pediatric high-grade gliomas and from those of adult high-grade gliomas. The molecular and clinical characteristics of DIPGs align with those of other midline high-grade gliomas with a specific H3 K27M mutation in histone H3.1 (H3F3A) or H3.3 (HIST1H3B and HIST1H3C), which led the World Health Organization to group these tumors together into a single entity.[1] In one report of 64 children with thalamic tumors, 50% of high-grade gliomas (11 of 22) had an H3 K27M mutation, and approximately 10% of tumors with low-grade morphological characteristics (5 of 42) had an H3 K27M mutation.[40] Five-year overall survival (OS) was only 6% (1 of 16). In another study that included 202 children with glioblastoma, 68 of the tumors were midline (primarily thalamic) and had an H3 K27M mutation.[35] Five-year OS for this group was only 5%, which was significantly inferior to the survival rates of the remaining patients in the study. A number of chromosomal and genomic abnormalities have been reported for DIPG, including the following: - Histone H3 genes: Approximately 80% of DIPG tumors have a mutation in a specific amino acid in the histone H3.1 (H3F3A) or H3.3 (HIST1H3B and HIST1H3C) genes.[33,41,42,43,44] This H3 K27M mutation is observed in pediatric high-grade gliomas at other midline locations but is uncommon in cortical pediatric high-grade gliomas and in adult high-grade gliomas.[32,33,41,42,43,44] An autopsy study that examined multiple tumor sites (primary, contiguous, and metastatic) in seven DIPG patients found that the H3 K27M mutation was invariably present, supporting its role as a driver mutation for DIPG.[45]
- Activin A receptor, type I (ACVR1) gene: Approximately 20% of DIPG cases have activating mutations in the ACVR1 gene, with most occurring concurrently with H3.3 mutations.[41,42,43,44] Germline mutations in ACVR1 cause the autosomal dominant syndrome fibrodysplasia ossificans progressiva (FOP), although there is no cancer predisposition in FOP.[46]
- Receptor tyrosine kinase amplification: PDGFRA amplification occurs in approximately 30% of cases, with lower rates of amplification observed for some other receptor tyrosine kinases (e.g., MET and IGF1R).[47,48] An autopsy study that examined multiple tumor sites (primary, contiguous, and metastatic) in seven DIPG patients found that PDGFRA amplification was variably present across these sites, suggesting that this change is a secondary genomic alteration in DIPG.[45]
- TP53 deletion: DIPG tumors commonly show deletion of the TP53 gene on chromosome 17p.[48] Additionally, TP53 is commonly mutated in DIPG tumors, particularly those with histone H3 gene mutations.[38,41,42,43,44] Aneuploidy is commonly observed in cases with TP53 mutations.[44]
The gene expression profile of DIPG differs from that of non-brain stem pediatric high-grade gliomas, further supporting a distinctive biology for this subset of pediatric gliomas.[48] Pediatric H3 K27M-mutant tumors rarely show MGMT promoter methylation,[35] which explains the lack of efficacy of temozolomide when it was tested in patients with DIPG.[49] (Refer to the PDQ summary on Childhood Brain Stem Glioma Treatment for information about the treatment of childhood brain stem gliomas.) Central Nervous System (CNS) Atypical Teratoid/Rhabdoid Tumors (AT/RT) SMARCB1gene AT/RT was the first primary pediatric brain tumor for which a candidate tumor suppressor gene, SMARCB1 (also known as INI1 and hSNF5), was identified.[50]SMARCB1 is genomically altered in the majority of rhabdoid tumors, including CNS, renal, and extrarenal rhabdoid malignancies.[50] Additional genomic alterations (mutations and gains/losses) in other genes are very uncommon in patients with SMARCB1-associated AT/RT, and there are no other genes that are recurrently mutated.[51,52,53] SMARCB1 is a component of a switch (SWI) and sucrose non-fermenting (SNF) adenosine triphosphate-dependent chromatin-remodeling complex.[54] Rare familial cases of rhabdoid tumors expressing SMARCB1 and lacking SMARCB1 mutations have also been associated with germline mutations of SMARCA4/BRG1, another member of the SWI/SNF chromatin-remodeling complex.[55,56] The 2016 WHO classification defines AT/RT by the presence of either SMARCB1 or SMARCA4 alterations. Tumors with histological features of AT/RT that lack these genomic alterations are termed CNS embryonal tumor with rhabdoid features.[1] Despite the absence of recurring genomic alterations beyond SMARCB1 (and, more rarely, other SWI/SNF complex members), biologically distinctive subsets of AT/RT have been identified.[57,58] The following three distinctive subsets of AT/RT were identified through the use of DNA methylation arrays for 150 AT/RT tumors and gene expression arrays for 67 AT/RT tumors:[58] - AT/RT TYR: This subset represented approximately one-third of cases and was characterized by elevated expression of melanosomal markers such as TYR (the gene encoding tyrosinase). Cases in this subset were primarily infratentorial, with most presenting in children aged 0 to 1 years and showing chromosome 22q loss.[58] For patients with AT/RT TYR, the mean overall survival (OS) is 37 months (95% confidence interval [CI], 18-56 months).[59] Cribriform neuroepithelial tumor is a brain cancer that also presents in young children and has genomic and epigenomic characteristics that are very similar to AT/RT TYR.[59]
- AT/RT SHH: This subset represented approximately 40% of cases and was characterized by elevated expression of genes in the sonic hedgehog (SHH) pathway (e.g., GLI2 and MYCN). Cases in this subset occurred with similar frequency in the supratentorium and infratentorium. While most presented before age 2 years, approximately one-third of cases presented between ages 2 and 5 years.[58] For patients with AT/RT SHH, the mean OS is 16 months (95% CI, 8-25 months).[59]
- AT/RT MYC: This subset represented approximately one-fourth of cases and was characterized by elevated expression of MYC. AT/RT MYC cases tended to occur in the supratentorial compartment. While most AT/RT MYC cases occurred by age 5 years, AT/RT MYC represented the most common subset diagnosed at age 6 years and older. Focal deletions of SMARCB1 were the most common mechanism of SMARCB1 loss for this subset.[58] For patients with AT/RT MYC, the mean OS is 13 months (95% CI, 5-22 months).[59]
In addition to somatic mutations, germline mutations in SMARCB1 have been reported in a substantial subset of AT/RT patients.[50,60] A study of 65 children with rhabdoid tumors found that 23 (35%) had germline mutations and/or deletions of SMARCB1.[61] Children with germline alterations in SMARCB1 presented at an earlier age than did sporadic cases (median age, approximately 5 months vs. 18 months) and were more likely to present with multiple tumors.[61] One parent was found to be a carrier of the SMARCB1 germline abnormality in 7 of 22 evaluated cases showing germline alterations, with four of the carrier parents being unaffected by SMARCB1-associated cancers.[61] This indicates that AT/RT shows an autosomal dominant inheritance pattern with incomplete penetrance. Gonadal mosaicism has also been observed, as evidenced by families in which multiple siblings are affected by AT/RT and have identical SMARCB1 alterations, but both parents lack a SMARCB1 mutation/deletion.[61,62] Screening children diagnosed with AT/RT for germline SMARCB1 mutations may provide useful information for counseling families on the genetic implications of their child's AT/RT diagnosis.[61] (Refer to the PDQ summary on Childhood Central Nervous System Atypical Teratoid/Rhabdoid Tumors Treatment for information about the treatment of childhood CNS atypical teratoid/rhabdoid tumors.) Medulloblastomas Multiple medulloblastoma subtypes have been identified by integrative molecular analysis.[63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78] Since 2012, the general consensus is that medulloblastoma can be molecularly separated into at least four core subtypes. These subtypes remain stable across primary and metastatic components.[79] It is likely that further subclassification will occur.[76,77,80] The 2016 World Health Organization (WHO) classification has endorsed this consensus by adding the following categories for genetically defined medulloblastoma:[1] - Medulloblastoma, WNT-activated.
- Medulloblastoma, sonic hedgehog (SHH)-activated and TP53-mutant.
- Medulloblastoma, SHH-activated and TP53-wildtype.
- Medulloblastoma, non-WNT/non-SHH.
The following four core molecularly defined subtypes of medulloblastoma have been identified:[76,77,81,82] - WNT medulloblastoma: WNT tumors are medulloblastomas with aberrations in the WNT signaling pathway. WNT medulloblastoma shows a WNT signaling gene expression signature and beta-catenin nuclear staining. They are usually histologically classified as classic medulloblastoma tumors and rarely have a large cell/anaplastic appearance. They are infrequently metastasized at diagnosis. Genetically, these tumors have 6q loss (monosomy 6), CTNNB1 mutations, and activated WNT signaling; MYC overexpression may be seen occasionally.[83]
The WNT subset is primarily observed in older children, adolescents, and adults and does not show a male predominance. The subset is believed to have brain stem origin, from the embryonal rhombic lip region. WNT medulloblastomas are associated with a very good outcome, especially in individuals whose tumors have beta-catenin nuclear staining and proven 6q loss and/or CTNNB1 mutations.[78,84] - SHH medulloblastoma: SHH tumors are medulloblastomas with aberrations in the SHH pathway. SHH medulloblastomas are characterized by chromosome 9q deletions; desmoplastic/nodular histology; and mutations in SHH pathway genes, including PTCH1, PTCH2, SMO, SUFU, and GLI2.
SHH medulloblastomas show a bimodal age distribution and are observed primarily in children younger than 3 years and in older adolescence/adulthood. The tumors are believed to emanate from the external granular layer of the cerebellum. Prognosis for patients with SHH medulloblastoma appears to be negatively affected by other molecular genetic changes, such as chromosome 17p loss, chromosome 3q gain, chromothripsis, p53 amplification, TP53 mutation, and the finding of large cell/anaplastic histology.[77,85] The outcome for patients with SHH medulloblastoma is relatively favorable, primarily in children younger than 3 years and in adults. This is likely because of the type of mutation present in the SHH pathway, given that patients with mutations upstream of the SHH signaling pathway, such as PTCH1, PTCH2, and SUFU, have a more favorable prognosis than do patients with downstream genomic aberrations, such as GLI2 and MYCN amplification.[86,87] Overall outcome in adolescents and young adults with SHH medulloblastoma is not different from that seen in patients with non-WNT pathway-activated tumors, except for patients with TP53 mutations and downstream SHH pathway mutations. Patients with unfavorable molecular findings have an unfavorable prognosis, with less than 50% of patients surviving after conventional treatment.[81,85,86,87,88] The 2016 WHO classification identifies SHH medulloblastoma with a TP53 mutation as a distinctive entity (medulloblastoma, SHH-activated and TP53-mutant).[1] Approximately 25% of SHH-activated medulloblastoma cases have TP53 mutations, with a high percentage of these cases also showing a TP53 germline mutation (9 of 20 in one study). These patients are commonly between the ages of 5 years and 18 years and have an inferior outcome (overall survival at 5 years, <50%).[85] The tumors often show large cell anaplastic histology.[85] - Group 3 medulloblastoma: Histology of group 3 medulloblastoma is either classic or large cell/anaplastic; these tumors are frequently metastasized at the time of diagnosis. A variety of different genomic aberrations have been noted in these tumors, including the presence of i17q and, most characteristically, MYC amplification.
Group 3 medulloblastomas occur throughout childhood and may occur in infants. Males outnumber females in a 2:1 ratio in this medulloblastoma subtype. Patients with group 3 medulloblastomas have a variable prognosis. Patients with MYC amplification or MYC overexpression have a poor prognosis, with less than 50% of these patients surviving 5 years after diagnosis. This poor prognosis is especially true in children younger than 4 years at diagnosis.[81] However, patients with group 3 medulloblastoma without MYC amplification or MYC overexpression who are older than 3 years have a prognosis similar to that of most patients with medulloblastoma, with a 5-year progression-free survival (PFS) rate higher than 70%.[88] - Group 4 medulloblastoma: Group 4 medulloblastomas are either classic or large cell/anaplastic tumors. Metastasis at diagnosis is common, but not as frequent as is seen in group 3 medulloblastomas. Molecularly, the medulloblastomas can have a CDK6 amplification, MYCN amplification, and most characteristically, an i17q abnormality.
Group 4 medulloblastomas occur throughout infancy and childhood and into adulthood. They also predominate in males. The prognosis is better than group 3 medulloblastoma but not as good as WNT medulloblastoma. Prognosis for group 4 medulloblastoma patients is affected by additional factors such as the presence of metastatic disease and chromosome 17p loss.[76,77]
Optimal ways of identifying the four core medulloblastoma subtypes for clinical use is under active study, and both immunohistochemical methods and methods based on gene expression analysis are under development.[84,89] The classification of medulloblastoma into the four major subtypes will be altered in the future.[90,91] Further subdivision within subgroups based on molecular characteristics is likely as each of the subgroups is further molecularly dissected, although there is no consensus regarding an alternative classification.[76,80,87] Whether the classification for adults with medulloblastoma has similar predictive ability in children is unknown.[77,81] In one study of adult medulloblastoma, MYC oncogene amplifications were rarely observed, and tumors with 6q deletion and WNT activation (as identified by nuclear beta-catenin staining) did not share the excellent prognosis seen in pediatric medulloblastomas, although another study did confirm an excellent prognosis for WNT-activated tumors in adults.[77,81] (Refer to the PDQ summary on Childhood Central Nervous System Embryonal Tumors Treatment for information about the treatment of childhood medulloblastoma.) Nonmedulloblastoma Embryonal Tumors This section describes the genomic characteristics of embryonal tumors other than medulloblastoma and atypical teratoid/rhabdoid tumor. The 2016 WHO classification removed the term primitive neuroectodermal tumors (PNET) from the diagnostic lexicon.[1] This change resulted from the recognition that many tumors previously classified as CNS PNETs have the common finding of amplification of the C19MC region on chromosome 19. These entities included ependymoblastoma, embryonal tumors with abundant neuropil and true rosettes (ETANTR), and some cases of medulloepithelioma. The 2016 WHO classification now categorizes tumors with C19MC amplification as embryonal tumor with multilayered rosettes (ETMR), C19MC-altered. Tumors previously classified as CNS PNETs are now termed CNS embryonal tumor, NOS, with the recognition that tumors in this category will likely be classified by their defining genomic lesions in future editions of the WHO classification. A study applying unsupervised clustering of DNA methylation patterns for 323 nonmedulloblastoma embryonal tumors found that approximately one-half of these tumors diagnosed as nonmedulloblastoma embryonal tumors showed molecular profiles characteristic of other known pediatric brain tumors (e.g., high-grade glioma, atypical teratoid/rhabdoid tumor).[92] This observation highlights the utility of molecular characterization to assign this class of tumors to their appropriate biology-based diagnosis. Among the same collection of 323 tumors diagnosed as nonmedulloblastoma embryonal tumors, molecular characterization identified genomically and biologically distinctive subtypes, including the following: - Embryonal tumors with multilayered rosettes (ETMR): Representing 11% of the 323 cases, this subtype combines embryonal rosette-forming neuroepithelial brain tumors that were previously categorized as either embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, or medulloepithelioma.[92,93] ETMRs arise in young children (median age at diagnosis, 2-3 years) and show a highly aggressive clinical course, with a median PFS of less than 1 year and few long-term survivors.[93]
ETMRs are defined at the molecular level by high-level amplification of the microRNA cluster C19MC and by a gene fusion between TTYH1 and C19MC.[93,94,95] This gene fusion puts expression of C19MC under control of the TTYH1 promoter, leading to high-level aberrant expression of the microRNAs within the cluster. The World Health Organization (WHO) allows histologically similar tumors without C19MC alteration to be classified as ETMR. - CNS neuroblastoma with FOXR2 activation (CNS NB-FOXR2): Representing 14% of the 323 cases, this subtype is characterized by genomic alterations that lead to increased expression of the transcription factor FOXR2.[92] CNS NB-FOXR2 is primarily observed in children younger than 10 years, and the histology of these tumors is typically that of CNS neuroblastoma or CNS ganglioneuroblastoma .[92] There is no single genomic alteration among CNS NB-FOXR2 tumors leading to FOXR2 overexpression, with gene fusions involving multiple FOXR2 partners identified.[92] This subtype has not been added to the WHO diagnostic lexicon.
- CNS Ewing sarcoma family tumor with CIC alteration (CNS EFT-CIC): Representing 4% of the 323 cases, this subtype is characterized by genomic alterations affecting CIC (located on chromosome 19q13.2), with fusion to NUTM1 being identified in several cases tested.[92]CIC gene fusions are also identified in extra-CNS Ewing-like sarcomas, and the gene expression signature of CNS EFT-CIC tumors is similar to that of these sarcomas.[92] CNS EFT-CIC tumors generally occur in children younger than 10 years and are characterized by a small cell phenotype but with variable histology.[92] This subtype has not been added to the WHO diagnostic lexicon.
- CNS high-grade neuroepithelial tumor with MN1 alteration (CNS HGNET-MN1): Representing 3% of the 323 cases, this subtype is characterized by gene fusions involving MN1 (located on chromosome 22q12.3), with fusion partners including BEND2 and CXXC5.[92] This subtype shows a striking female predominance and tends to occur in the second decade of life.[92] This subtype contained most cases carrying a diagnosis of astroblastoma as per the 2007 WHO classification scheme.[92] This subtype has not been added to the WHO diagnostic lexicon.
- CNS high-grade neuroepithelial tumor with BCOR alteration (CNS HGNET-BCOR): Representing 3% of the 323 cases, this subtype is characterized by internal tandem duplications of BCOR,[92] a genomic alteration that is also found in clear cell sarcoma of the kidney.[96,97] While the median age at diagnosis is younger than 10 years, cases arising in the second decade of life and beyond do occur.[92] This subtype has not been added to the WHO diagnostic lexicon.
Medulloepithelioma Medulloepithelioma is identified as a histologically discrete tumor within the WHO classification system.[98,99] Medulloepithelioma tumors are rare and tend to arise most commonly in infants and young children. Medulloepitheliomas, which histologically recapitulate the embryonal neural tube, tend to arise supratentorially, primarily intraventricularly, but may arise infratentorially, in the cauda, and even extraneurally, along nerve roots.[98,99] Medulloepithelioma with the classic molecular change is considered an ETMR. Pineoblastoma Pineoblastoma, which was previously conventionally grouped with embryonal tumors, is now categorized by the WHO as a pineal parenchymal tumor. Given that therapies for pineoblastoma are quite similar to those utilized for embryonal tumors, the previous convention of including pineoblastoma with the CNS embryonal tumors is followed here. Pineoblastoma is associated with germline mutations in both the retinoblastoma (RB1) gene and in DICER1, as described below: - Pineoblastoma is associated with germline mutations in RB1, with the term trilateral retinoblastoma used to refer to ocular retinoblastoma in combination with a histologically similar brain tumor generally arising in the pineal gland or other midline structures. Historically, intracranial tumors have been reported in 5% to 15% of children with heritable retinoblastoma.[100] Rates of pineoblastoma among children with heritable retinoblastoma who undergo current treatment programs may be lower than these historical estimates.[101,102,103]
- Germline DICER1 mutations have also been reported in patients with pineoblastoma.[104] Among 18 patients with pineoblastoma, three patients with DICER1 germline mutations were identified, and an additional three patients known to be carriers of germline DICER1 mutations developed pineoblastoma.[104] The DICER1 mutations in patients with pineoblastoma are loss-of-function mutations that appear to be distinct from the mutations observed in DICER1 syndrome-related tumors such as pleuropulmonary blastoma.[104]
(Refer to the PDQ summary on Childhood Central Nervous System Embryonal Tumors Treatment for information about the treatment of childhood PNETs.) Ependymomas Molecular characterization studies have identified several biological subtypes of ependymoma based on their distinctive DNA methylation and gene expression profiles and on their distinctive spectrum of genomic alterations.[105,106,107] - Infratentorial tumors.
- Posterior fossa A, CpG island methylator phenotype (CIMP)-positive ependymoma, termed EPN-PFA.
- Posterior fossa B, CIMP-negative ependymoma, termed EPN-PFB.
- Supratentorial tumors.
- C11orf95-RELA-positive ependymoma.
- C11orf95-RELA-negative and YAP1 fusion-positive ependymoma.
- Spinal tumors.
Figure 6. Graphical summary of key molecular and clinical characteristics of ependymal tumor subgroups.Schematic representation of key genetic and epigenetic findings in the nine molecular subgroups of ependymal tumors as identified by methylation profiling. CIN, Chromosomal instability. Reprinted from Cancer Cell, Volume 27, Kristian W. Pajtler, Hendrik Witt, Martin Sill, David T.W. Jones, Volker Hovestadt, Fabian Kratochwil, Khalida Wani, Ruth Tatevossian, Chandanamali Punchihewa, Pascal Johann, Juri Reimand, Hans-Jorg Warnatz, Marina Ryzhova, Steve Mack, Vijay Ramaswamy, David Capper, Leonille Schweizer, Laura Sieber, Andrea Wittmann, Zhiqin Huang, Peter van Sluis, Richard Volckmann, Jan Koster, Rogier Versteeg, Daniel Fults, Helen Toledano, Smadar Avigad, Lindsey M. Hoffman, Andrew M. Donson, Nicholas Foreman, Ekkehard Hewer, Karel Zitterbart, Mark Gilbert, Terri S. Armstrong, Nalin Gupta, Jeffrey C. Allen, Matthias A. Karajannis, David Zagzag, Martin Hasselblatt, Andreas E. Kulozik, Olaf Witt, V. Peter Collins, Katja von Hoff, Stefan Rutkowski, Torsten Pietsch, Gary Bader, Marie-Laure Yaspo, Andreas von Deimling, Peter Lichter, Michael D. Taylor, Richard Gilbertson, David W. Ellison, Kenneth Aldape, Andrey Korshunov, Marcel Kool, and Stefan M. Pfister, Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups, Pages 728-743, Copyright (2015), with permission from Elsevier.
Approximately two-thirds of childhood ependymomas arise in the posterior fossa, and two major genomically defined subtypes of posterior fossa tumors are recognized. Similarly, most pediatric supratentorial tumors can be categorized into one of two genomic subtypes. These subtypes and their associated clinical characteristics are described below.[105] Among these subtypes, the 2016 World Health Organization (WHO) classification has accepted ependymoma, RELA fusion-positive as a distinct diagnostic entity.[1] The most common posterior fossa ependymoma subtype is EPN-PFA and is characterized by the following: - Presentation in young children (median age, 3 years).[105]
- Low rates of mutations that affect protein structure (approximately five per genome), with no recurring mutations.[106]
- A balanced chromosomal profile (refer to Figure 7) with few chromosomal gains or losses.[105,106]
Figure 7. Identification of Subgroup-Specific Copy Number Alterations in the Posterior Fossa Ependymoma Genome. (A) Copy number profiling of 75 PF ependymomas using 10K array-CGH identifies disparate genetic landscapes between Group A and Group B tumors. Toronto and Heidelberg copy number datasets have been combined and summarized in a heatmap. The heatmap also displays the association of tumors to cytogenetic risk groups 1, 2, and 3 (Korshunov et al., 2010). Statistically significant chromosomal aberrations (black boxes) are also displayed between both subgroups, calculated by Fisher's exact test. Witt H, Mack SC, Ryzhova M, et al.: Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20 (2): 143-57, 2011, doi:10.1016/j.ccr.2011.07.007. Copyright © 2011 Elsevier Inc. All rights reserved.
- Gain of chromosome 1q, a known poor prognostic factor for ependymomas,[108] in approximately 25% of cases.[105,107]
- Presence of the CIMP (i.e., CIMP positive).[107]
- High rates of disease recurrence (33% progression-free survival [PFS] at 5 years) and low survival rates compared with other subtypes (68% at 5 years).[105]
The EPN-PFB subtype is less common than the EPN-PFA subtype in children and is characterized by the following: - Presentation primarily in adolescents and young adults (median age, 30 years).[105]
- Low rates of mutations that affect protein structure (approximately five per genome), with no recurring mutations.[107]
- Numerous cytogenetic abnormalities (refer to Figure 7), primarily involving the gain/loss of whole chromosomes.[105,107]
- Absence of the CIMP (i.e., CIMP negative).[107]
- Favorable outcome in comparison to EPN-PFA, with 5-year PFS of 73% and overall survival (OS) of 100%.[105]
The largest subset of pediatric supratentorial (ST) ependymomas are characterized by gene fusions involving RELA,[109,110] a transcriptional factor important in NF-κB pathway activity. This subtype is termed ST-EPN-RELA and is characterized by the following: - Represents approximately 70% of supratentorial ependymomas in children,[109,110] and presents at a median age of 8 years.[105]
- Presence of C11orf95-RELA fusions resulting from chromothripsis involving chromosome 11q13.1.[109]
- Evidence of NF-κB pathway activation at the protein and RNA level.[109]
- Low rates of mutations that affect protein structure and absence of recurring mutations outside of C11orf95-RELA fusions.[109]
- Presence of homozygous deletions of CDKN2A, a known poor prognostic factor for ependymomas,[108] in approximately 15% of cases.[105]
- Gain of chromosome 1q, a known poor prognostic factor for ependymomas, in approximately one-quarter of cases.[105]
- Unfavorable outcome in comparison to other ependymoma subtypes, with 5-year PFS of 29% and OS of 75%.[105]
- Supratentorial clear cell ependymomas with branching capillaries commonly show the C11orf95-RELA fusion,[111] and one series of 20 patients with a median age of 10.4 years showed a relatively favorable prognosis (5-year PFS of 68% and OS of 72%).[111]
A second, less common subset of supratentorial ependymomas, termed ST-EPN-YAP1, has fusions involving YAP1 and are characterized by the following: - Median age at diagnosis of 1.4 years.[105]
- Presence of a gene fusion involving YAP1, with MAMLD1 being the most common fusion partner.[105,109]
- A relatively stable genome with few chromosomal changes other than the YAP1 fusion.[105]
- Relatively favorable prognosis (although based on small numbers), with a 5-year PFS of 66% and OS of 100%.[105]
Clinical implications of genomic alterations The absence of recurring mutations in the EPN-PFA and EPN-PFB subtypes at diagnosis precludes using their genomic profiles to guide therapy. The RELA and YAP1 fusion genes present in supratentorial ependymomas are not directly targetable with agents in the clinic, but can provide leads for future research. (Refer to the PDQ summary on Childhood Ependymoma Treatment for information about the treatment of childhood ependymoma.) References:
-
Louis DN, Perry A, Reifenberger G, et al.: The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131 (6): 803-20, 2016.
-
Bar EE, Lin A, Tihan T, et al.: Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol 67 (9): 878-87, 2008.
-
Forshew T, Tatevossian RG, Lawson AR, et al.: Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol 218 (2): 172-81, 2009.
-
Jones DT, Kocialkowski S, Liu L, et al.: Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68 (21): 8673-7, 2008.
-
Jones DT, Kocialkowski S, Liu L, et al.: Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene 28 (20): 2119-23, 2009.
-
Pfister S, Janzarik WG, Remke M, et al.: BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest 118 (5): 1739-49, 2008.
-
Korshunov A, Meyer J, Capper D, et al.: Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol 118 (3): 401-5, 2009.
-
Horbinski C, Hamilton RL, Nikiforov Y, et al.: Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathol 119 (5): 641-9, 2010.
-
Yu J, Deshmukh H, Gutmann RJ, et al.: Alterations of BRAF and HIPK2 loci predominate in sporadic pilocytic astrocytoma. Neurology 73 (19): 1526-31, 2009.
-
Lin A, Rodriguez FJ, Karajannis MA, et al.: BRAF alterations in primary glial and glioneuronal neoplasms of the central nervous system with identification of 2 novel KIAA1549:BRAF fusion variants. J Neuropathol Exp Neurol 71 (1): 66-72, 2012.
-
Hawkins C, Walker E, Mohamed N, et al.: BRAF-KIAA1549 fusion predicts better clinical outcome in pediatric low-grade astrocytoma. Clin Cancer Res 17 (14): 4790-8, 2011.
-
Becker AP, Scapulatempo-Neto C, Carloni AC, et al.: KIAA1549: BRAF Gene Fusion and FGFR1 Hotspot Mutations Are Prognostic Factors in Pilocytic Astrocytomas. J Neuropathol Exp Neurol 74 (7): 743-54, 2015.
-
Horbinski C, Nikiforova MN, Hagenkord JM, et al.: Interplay among BRAF, p16, p53, and MIB1 in pediatric low-grade gliomas. Neuro Oncol 14 (6): 777-89, 2012.
-
Roth JJ, Fierst TM, Waanders AJ, et al.: Whole Chromosome 7 Gain Predicts Higher Risk of Recurrence in Pediatric Pilocytic Astrocytomas Independently From KIAA1549-BRAF Fusion Status. J Neuropathol Exp Neurol 75 (4): 306-15, 2016.
-
Mistry M, Zhukova N, Merico D, et al.: BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol 33 (9): 1015-22, 2015.
-
Janzarik WG, Kratz CP, Loges NT, et al.: Further evidence for a somatic KRAS mutation in a pilocytic astrocytoma. Neuropediatrics 38 (2): 61-3, 2007.
-
Dougherty MJ, Santi M, Brose MS, et al.: Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol 12 (7): 621-30, 2010.
-
Dias-Santagata D, Lam Q, Vernovsky K, et al.: BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: diagnostic and therapeutic implications. PLoS One 6 (3): e17948, 2011.
-
Schindler G, Capper D, Meyer J, et al.: Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol 121 (3): 397-405, 2011.
-
Dahiya S, Haydon DH, Alvarado D, et al.: BRAF(V600E) mutation is a negative prognosticator in pediatric ganglioglioma. Acta Neuropathol 125 (6): 901-10, 2013.
-
Ho CY, Mobley BC, Gordish-Dressman H, et al.: A clinicopathologic study of diencephalic pediatric low-grade gliomas with BRAF V600 mutation. Acta Neuropathol 130 (4): 575-85, 2015.
-
Bandopadhayay P, Ramkissoon LA, Jain P, et al.: MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet 48 (3): 273-82, 2016.
-
Jones DT, Hutter B, Jäger N, et al.: Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 45 (8): 927-32, 2013.
-
Zhang J, Wu G, Miller CP, et al.: Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45 (6): 602-12, 2013.
-
Ramkissoon LA, Horowitz PM, Craig JM, et al.: Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci U S A 110 (20): 8188-93, 2013.
-
Franz DN, Belousova E, Sparagana S, et al.: Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 381 (9861): 125-32, 2013.
-
Qaddoumi I, Orisme W, Wen J, et al.: Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol 131 (6): 833-45, 2016.
-
Pollack IF, Hamilton RL, Sobol RW, et al.: IDH1 mutations are common in malignant gliomas arising in adolescents: a report from the Children's Oncology Group. Childs Nerv Syst 27 (1): 87-94, 2011.
-
Paugh BS, Qu C, Jones C, et al.: Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol 28 (18): 3061-8, 2010.
-
Bax DA, Mackay A, Little SE, et al.: A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res 16 (13): 3368-77, 2010.
-
Ward SJ, Karakoula K, Phipps KP, et al.: Cytogenetic analysis of paediatric astrocytoma using comparative genomic hybridisation and fluorescence in-situ hybridisation. J Neurooncol 98 (3): 305-18, 2010.
-
Schwartzentruber J, Korshunov A, Liu XY, et al.: Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482 (7384): 226-31, 2012.
-
Wu G, Broniscer A, McEachron TA, et al.: Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44 (3): 251-3, 2012.
-
Sturm D, Witt H, Hovestadt V, et al.: Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22 (4): 425-37, 2012.
-
Korshunov A, Ryzhova M, Hovestadt V, et al.: Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol 129 (5): 669-78, 2015.
-
Aihara K, Mukasa A, Gotoh K, et al.: H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro Oncol 16 (1): 140-6, 2014.
-
Gielen GH, Gessi M, Hammes J, et al.: H3F3A K27M mutation in pediatric CNS tumors: a marker for diffuse high-grade astrocytomas. Am J Clin Pathol 139 (3): 345-9, 2013.
-
Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al.: K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 124 (3): 439-47, 2012.
-
Gielen GH, Gessi M, Buttarelli FR, et al.: Genetic Analysis of Diffuse High-Grade Astrocytomas in Infancy Defines a Novel Molecular Entity. Brain Pathol 25 (4): 409-17, 2015.
-
Ryall S, Krishnatry R, Arnoldo A, et al.: Targeted detection of genetic alterations reveal the prognostic impact of H3K27M and MAPK pathway aberrations in paediatric thalamic glioma. Acta Neuropathol Commun 4 (1): 93, 2016.
-
Wu G, Diaz AK, Paugh BS, et al.: The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46 (5): 444-50, 2014.
-
Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, et al.: Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 46 (5): 462-6, 2014.
-
Taylor KR, Mackay A, Truffaux N, et al.: Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 46 (5): 457-61, 2014.
-
Buczkowicz P, Hoeman C, Rakopoulos P, et al.: Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet 46 (5): 451-6, 2014.
-
Hoffman LM, DeWire M, Ryall S, et al.: Spatial genomic heterogeneity in diffuse intrinsic pontine and midline high-grade glioma: implications for diagnostic biopsy and targeted therapeutics. Acta Neuropathol Commun 4: 1, 2016.
-
Shore EM, Xu M, Feldman GJ, et al.: A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet 38 (5): 525-7, 2006.
-
Zarghooni M, Bartels U, Lee E, et al.: Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol 28 (8): 1337-44, 2010.
-
Paugh BS, Broniscer A, Qu C, et al.: Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol 29 (30): 3999-4006, 2011.
-
Cohen KJ, Heideman RL, Zhou T, et al.: Temozolomide in the treatment of children with newly diagnosed diffuse intrinsic pontine gliomas: a report from the Children's Oncology Group. Neuro Oncol 13 (4): 410-6, 2011.
-
Biegel JA, Tan L, Zhang F, et al.: Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res 8 (11): 3461-7, 2002.
-
Lee RS, Stewart C, Carter SL, et al.: A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest 122 (8): 2983-8, 2012.
-
Kieran MW, Roberts CW, Chi SN, et al.: Absence of oncogenic canonical pathway mutations in aggressive pediatric rhabdoid tumors. Pediatr Blood Cancer 59 (7): 1155-7, 2012.
-
Hasselblatt M, Isken S, Linge A, et al.: High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes Chromosomes Cancer 52 (2): 185-90, 2013.
-
Biegel JA, Kalpana G, Knudsen ES, et al.: The role of INI1 and the SWI/SNF complex in the development of rhabdoid tumors: meeting summary from the workshop on childhood atypical teratoid/rhabdoid tumors. Cancer Res 62 (1): 323-8, 2002.
-
Schneppenheim R, Frühwald MC, Gesk S, et al.: Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet 86 (2): 279-84, 2010.
-
Hasselblatt M, Gesk S, Oyen F, et al.: Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am J Surg Pathol 35 (6): 933-5, 2011.
-
Torchia J, Picard D, Lafay-Cousin L, et al.: Molecular subgroups of atypical teratoid rhabdoid tumours in children: an integrated genomic and clinicopathological analysis. Lancet Oncol 16 (5): 569-82, 2015.
-
Johann PD, Erkek S, Zapatka M, et al.: Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell 29 (3): 379-93, 2016.
-
Johann PD, Hovestadt V, Thomas C, et al.: Cribriform neuroepithelial tumor: Molecular characterization of a SMARCB1-deficient non-rhabdoid tumor with favorable long-term outcome. Brain Pathol : , 2016.
-
Biegel JA, Fogelgren B, Wainwright LM, et al.: Germline INI1 mutation in a patient with a central nervous system atypical teratoid tumor and renal rhabdoid tumor. Genes Chromosomes Cancer 28 (1): 31-7, 2000.
-
Eaton KW, Tooke LS, Wainwright LM, et al.: Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer 56 (1): 7-15, 2011.
-
Bruggers CS, Bleyl SB, Pysher T, et al.: Clinicopathologic comparison of familial versus sporadic atypical teratoid/rhabdoid tumors (AT/RT) of the central nervous system. Pediatr Blood Cancer 56 (7): 1026-31, 2011.
-
Onvani S, Etame AB, Smith CA, et al.: Genetics of medulloblastoma: clues for novel therapies. Expert Rev Neurother 10 (5): 811-23, 2010.
-
Dubuc AM, Northcott PA, Mack S, et al.: The genetics of pediatric brain tumors. Curr Neurol Neurosci Rep 10 (3): 215-23, 2010.
-
Thompson MC, Fuller C, Hogg TL, et al.: Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol 24 (12): 1924-31, 2006.
-
Kool M, Koster J, Bunt J, et al.: Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 3 (8): e3088, 2008.
-
Tabori U, Baskin B, Shago M, et al.: Universal poor survival in children with medulloblastoma harboring somatic TP53 mutations. J Clin Oncol 28 (8): 1345-50, 2010.
-
Pfister S, Remke M, Benner A, et al.: Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol 27 (10): 1627-36, 2009.
-
Ellison DW, Onilude OE, Lindsey JC, et al.: beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children's Cancer Study Group Brain Tumour Committee. J Clin Oncol 23 (31): 7951-7, 2005.
-
Polkinghorn WR, Tarbell NJ: Medulloblastoma: tumorigenesis, current clinical paradigm, and efforts to improve risk stratification. Nat Clin Pract Oncol 4 (5): 295-304, 2007.
-
Giangaspero F, Wellek S, Masuoka J, et al.: Stratification of medulloblastoma on the basis of histopathological grading. Acta Neuropathol 112 (1): 5-12, 2006.
-
Northcott PA, Korshunov A, Witt H, et al.: Medulloblastoma comprises four distinct molecular variants. J Clin Oncol 29 (11): 1408-14, 2011.
-
Pomeroy SL, Tamayo P, Gaasenbeek M, et al.: Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415 (6870): 436-42, 2002.
-
Jones DT, Jäger N, Kool M, et al.: Dissecting the genomic complexity underlying medulloblastoma. Nature 488 (7409): 100-5, 2012.
-
Peyrl A, Chocholous M, Kieran MW, et al.: Antiangiogenic metronomic therapy for children with recurrent embryonal brain tumors. Pediatr Blood Cancer 59 (3): 511-7, 2012.
-
Taylor MD, Northcott PA, Korshunov A, et al.: Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123 (4): 465-72, 2012.
-
Kool M, Korshunov A, Remke M, et al.: Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123 (4): 473-84, 2012.
-
Pietsch T, Schmidt R, Remke M, et al.: Prognostic significance of clinical, histopathological, and molecular characteristics of medulloblastomas in the prospective HIT2000 multicenter clinical trial cohort. Acta Neuropathol 128 (1): 137-49, 2014.
-
Wang X, Dubuc AM, Ramaswamy V, et al.: Medulloblastoma subgroups remain stable across primary and metastatic compartments. Acta Neuropathol 129 (3): 449-57, 2015.
-
Northcott PA, Jones DT, Kool M, et al.: Medulloblastomics: the end of the beginning. Nat Rev Cancer 12 (12): 818-34, 2012.
-
Cho YJ, Tsherniak A, Tamayo P, et al.: Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 29 (11): 1424-30, 2011.
-
Gajjar A, Bowers DC, Karajannis MA, et al.: Pediatric Brain Tumors: Innovative Genomic Information Is Transforming the Diagnostic and Clinical Landscape. J Clin Oncol 33 (27): 2986-98, 2015.
-
Ellison DW, Kocak M, Dalton J, et al.: Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol 29 (11): 1400-7, 2011.
-
Ellison DW, Dalton J, Kocak M, et al.: Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol 121 (3): 381-96, 2011.
-
Zhukova N, Ramaswamy V, Remke M, et al.: Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J Clin Oncol 31 (23): 2927-35, 2013.
-
Shih DJ, Northcott PA, Remke M, et al.: Cytogenetic prognostication within medulloblastoma subgroups. J Clin Oncol 32 (9): 886-96, 2014.
-
Kool M, Jones DT, Jäger N, et al.: Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25 (3): 393-405, 2014.
-
Schwalbe EC, Williamson D, Lindsey JC, et al.: DNA methylation profiling of medulloblastoma allows robust subclassification and improved outcome prediction using formalin-fixed biopsies. Acta Neuropathol 125 (3): 359-71, 2013.
-
Northcott PA, Shih DJ, Remke M, et al.: Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol 123 (4): 615-26, 2012.
-
Gottardo NG, Hansford JR, McGlade JP, et al.: Medulloblastoma Down Under 2013: a report from the third annual meeting of the International Medulloblastoma Working Group. Acta Neuropathol 127 (2): 189-201, 2014.
-
Louis DN, Perry A, Burger P, et al.: International Society Of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 24 (5): 429-35, 2014.
-
Sturm D, Orr BA, Toprak UH, et al.: New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 164 (5): 1060-72, 2016.
-
Korshunov A, Sturm D, Ryzhova M, et al.: Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity. Acta Neuropathol 128 (2): 279-89, 2014.
-
Kleinman CL, Gerges N, Papillon-Cavanagh S, et al.: Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet 46 (1): 39-44, 2014.
-
Li M, Lee KF, Lu Y, et al.: Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell 16 (6): 533-46, 2009.
-
Ueno-Yokohata H, Okita H, Nakasato K, et al.: Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat Genet 47 (8): 861-3, 2015.
-
Roy A, Kumar V, Zorman B, et al.: Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat Commun 6: 8891, 2015.
-
Louis DN, Ohgaki H, Wiestler OD, et al.: The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114 (2): 97-109, 2007.
-
Sharma MC, Mahapatra AK, Gaikwad S, et al.: Pigmented medulloepithelioma: report of a case and review of the literature. Childs Nerv Syst 14 (1-2): 74-8, 1998 Jan-Feb.
-
de Jong MC, Kors WA, de Graaf P, et al.: Trilateral retinoblastoma: a systematic review and meta-analysis. Lancet Oncol 15 (10): 1157-67, 2014.
-
Ramasubramanian A, Kytasty C, Meadows AT, et al.: Incidence of pineal gland cyst and pineoblastoma in children with retinoblastoma during the chemoreduction era. Am J Ophthalmol 156 (4): 825-9, 2013.
-
Abramson DH, Dunkel IJ, Marr BP, et al.: Incidence of pineal gland cyst and pineoblastoma in children with retinoblastoma during the chemoreduction era. Am J Ophthalmol 156 (6): 1319-20, 2013.
-
Turaka K, Shields CL, Meadows AT, et al.: Second malignant neoplasms following chemoreduction with carboplatin, etoposide, and vincristine in 245 patients with intraocular retinoblastoma. Pediatr Blood Cancer 59 (1): 121-5, 2012.
-
de Kock L, Sabbaghian N, Druker H, et al.: Germ-line and somatic DICER1 mutations in pineoblastoma. Acta Neuropathol 128 (4): 583-95, 2014.
-
Pajtler KW, Witt H, Sill M, et al.: Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 27 (5): 728-43, 2015.
-
Witt H, Mack SC, Ryzhova M, et al.: Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20 (2): 143-57, 2011.
-
Mack SC, Witt H, Piro RM, et al.: Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506 (7489): 445-50, 2014.
-
Korshunov A, Witt H, Hielscher T, et al.: Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol 28 (19): 3182-90, 2010.
-
Parker M, Mohankumar KM, Punchihewa C, et al.: C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature 506 (7489): 451-5, 2014.
-
Pietsch T, Wohlers I, Goschzik T, et al.: Supratentorial ependymomas of childhood carry C11orf95-RELA fusions leading to pathological activation of the NF-κB signaling pathway. Acta Neuropathol 127 (4): 609-11, 2014.
-
Figarella-Branger D, Lechapt-Zalcman E, Tabouret E, et al.: Supratentorial clear cell ependymomas with branching capillaries demonstrate characteristic clinicopathological features and pathological activation of nuclear factor-kappaB signaling. Neuro Oncol 18 (7): 919-27, 2016.
Hepatoblastoma and Hepatocellular CarcinomaGenomic abnormalities related to hepatoblastoma include the following: - Hepatoblastoma mutation frequency, as determined by three groups using whole-exome sequencing, was very low (approximately three variants per tumor) in children younger than 5 years.[1,2,3]
- Hepatoblastoma is primarily a disease of WNT pathway activation. The primary mechanism for WNT pathway activation is CTNNB1 activating mutations/deletions involving exon 3. CTNNB1 mutations have been reported in 70% of cases.[1] Rare causes of WNT pathway activation include mutations in AXIN1, AXIN2, and APC (APC seen only in cases associated with familial adenomatosis polyposis coli).[4]
- The frequency of NFE2L2 mutations in hepatoblastoma specimens was reported to be 4 (7%) of 62 tumors in one study [2] and 5 (10%) of 51 specimens in another study.[1] Similar mutations have been found in many types of cancer including hepatocellular carcinoma. These mutations render NFE2L2 insensitive to KEAP1-mediated degradation, leading to activation of the NFE2L2-KEAP1 pathway, which activates resistance to oxidative stress and is believed to confer resistance to chemotherapy.
- Somatic mutations were identified in other genes related to regulation of oxidative stress, including inactivating mutations in the thioredoxin-domain containing genes, TXNDC15 and TXNDC16.[2]
- Figure 8 shows the distribution of CTNNB1, NFE2L2, and TERT mutations for hepatoblastoma.[1]
Figure 8. Mutational status and functional relevance of NFE2L2 in hepatoblastoma. Clinicopathological characteristics and the mutational status of the CTNNB1, APC, and NFE2L2 genes, as well as the TERT promoter region are color-coded and depicted in rows for each tumor of our cohort of 43 hepatoblastoma (HB) patients and four transitional liver cell tumour (TLCT) patients and 4 HB cell lines. Reprinted from Journal of Hepatology, Volume 61 (Issue 6), Melanie Eichenmüller, Franziska Trippel, Michaela Kreuder, Alexander Beck, Thomas Schwarzmayr, Beate Häberle, Stefano Cairo, Ivo Leuschner, Dietrich von Schweinitz, Tim M. Strom, Roland Kappler, The genomic landscape of hepatoblastoma and their progenies with HCC-like features, Pages 1312-1320, Copyright 2014, with permission from Elsevier.
Genomic abnormalities related to hepatocellular carcinoma include the following: - A first case of pediatric hepatocellular carcinoma was analyzed by whole-exome sequencing, which showed a higher mutation rate (53 variants) and the coexistence of CTNNB1 and NFE2L2 mutations.[5]
- Fibrolamellar hepatocellular carcinoma, a rare subtype of hepatocellular carcinoma observed in older children, is characterized by an approximately 400 kB deletion on chromosome 19 that results in production of a chimeric RNA coding for a protein containing the amino-terminal domain of DNAJB1, a homolog of the molecular chaperone DNAJ, fused in frame with PRKACA, the catalytic domain of protein kinase A.[6]
- A rare, more aggressive subtype of childhood liver cancer (hepatocellular carcinoma, not otherwise specified, also termed transitional liver cell tumor) occurs in older children, and it has clinical and histopathological findings of both hepatoblastoma and hepatocellular carcinoma. TERT mutations were observed in two of four cases tested.[1]TERT mutations are also commonly observed in adults with hepatocellular carcinoma.[7]
(Refer to the PDQ summary on Childhood Liver Cancer Treatment for information about the treatment of liver cancer.) References:
-
Eichenmüller M, Trippel F, Kreuder M, et al.: The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol 61 (6): 1312-20, 2014.
-
Trevino LR, Wheeler DA, Finegold MJ, et al.: Exome sequencing of hepatoblastoma reveals recurrent mutations in NFE2L2. [Abstract] Cancer Res 73 (8 Suppl): A-4592, 2013. Also available online. Last accessed February 03, 2017.
-
Jia D, Dong R, Jing Y, et al.: Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology 60 (5): 1686-96, 2014.
-
Hiyama E, Kurihara S, Onitake Y: Integrated exome analysis in childhood hepatoblastoma: Biological approach for next clinical trial designs. [Abstract] Cancer Res 74 (19 Suppl): A-5188, 2014.
-
Vilarinho S, Erson-Omay EZ, Harmanci AS, et al.: Paediatric hepatocellular carcinoma due to somatic CTNNB1 and NFE2L2 mutations in the setting of inherited bi-allelic ABCB11 mutations. J Hepatol 61 (5): 1178-83, 2014.
-
Honeyman JN, Simon EP, Robine N, et al.: Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 343 (6174): 1010-4, 2014.
-
Nault JC, Mallet M, Pilati C, et al.: High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun 4: 2218, 2013.
SarcomasOsteosarcoma The genomic landscape of osteosarcoma is distinctive from that of other childhood cancers. It is characterized by an exceptionally high number of structural variants with relatively small numbers of single nucleotide variants in comparison to many adult cancers.[1,2] Key observations regarding the genomic landscape of osteosarcoma are summarized below: A number of germline mutations are associated with susceptibility to osteosarcoma; Table 1 summarizes the syndromes and associated genes for these conditions. Mutations in TP53 are the most common germline alterations associated with osteosarcoma. Mutations in this gene are found in approximately 70% of patients with Li-Fraumeni syndrome (LFS), which is associated with increased risk of osteosarcoma, breast cancer, various brain cancers, soft tissue sarcomas, and other cancers. While rhabdomyosarcoma is the most common sarcoma arising in patients aged 5 years and younger with TP53-associated LFS, osteosarcoma is the most common sarcoma in children and adolescents aged 6 to 19 years.[3] One study observed a high frequency of young osteosarcoma cases (age <30 years) carrying a known LFS- or likely LFS-associated TP53 mutation (3.8%) or rare exonic TP53 variant (5.7%), with an overall TP53 mutation frequency of 9.5%.[4] Another study observed germline mutations in TP53 in 7 of 59 (12%) osteosarcoma cases subjected to whole-exome sequencing.[2] Other groups have reported lower rates (3%-7%) of TP53 germline mutations in patients with osteosarcoma.[5,6] Table 1. Genetic Diseases That Predispose to Osteosarcomaa| Syndrome | Description | Location | Gene | Function |
|---|
| AML = acute myeloid leukemia; IL-1 = interleukin-1; MDS = myelodysplastic syndrome; RANK = receptor activator of nuclear factor kappaB ligand; TNF = tumor necrosis factor. | | a Table adapted from Kansara et al.[7] | | Bloom syndrome[8] | Rare inherited disorder characterized by short stature and sun-sensitive skin changes. Often presents with a long, narrow face, small lower jaw, large nose, and prominent ears. | 15q26.1 | BLM(RecQL3) | DNA helicase | | Diamond-Blackfan anemia[9] | Inherited pure red cell aplasia. Patients at risk for MDS and AML. Associated with skeletal abnormalities, such as abnormal facial features (flat nasal bridge, widely spaced eyes). | | Ribosomal proteins | Ribosome production[9,10] | | Li-Fraumeni syndrome[11] | Inherited mutation inTP53gene. Affected family members at increased risk for bone tumors, breast cancer, leukemia, brain tumors, and sarcomas. | 17p13.1 | P53 | DNA damage response | | Paget disease[12] | Excessive breakdown of bone with abnormal bone formation and remodeling, resulting in pain from weak, malformed bone. | 18q21-qa22 | LOH18CR1 | IL-1/TNF signaling; RANK signaling pathway | | 5q31 | | 5q35-qter | | Retinoblastoma[13] | Malignant tumor of the retina. Approximately 66% of patients diagnosed by age 2 years and 95% of patients by age 3 years. Patients with heritable germ cell mutations at greater risk for subsequent neoplasms. | 13q14.2 | RB1 | Cell-cycle checkpoint | | Rothmund-Thomson syndrome (also called poikiloderma congenitale)[14,15] | Autosomal recessive condition. Associated with skin findings (atrophy, telangiectasias, pigmentation), sparse hair, cataracts, small stature, and skeletal abnormalities. Increased incidence of osteosarcoma at a younger age. | 8q24.3 | RTS(RecQL4) | DNA helicase | | Werner syndrome[16] | Patients often have short stature and in their early twenties, develop signs of aging, including graying of hair and hardening of skin. Other aging problems such as cataracts, skin ulcers, and atherosclerosis develop later. | 8p12-p11.2 | WRN(RecQL2) | DNA helicase; exonuclease activity | Refer to the following summaries for more information about these genetic syndromes: - Genetics of Breast and Gynecologic Cancers (Li-Fraumeni syndrome).
- Genetics of Skin Cancer (Bloom syndrome, Rothmund-Thomson syndrome, and Werner syndrome).
(Refer to the PDQ summary on Osteosarcoma and Malignant Fibrous Histiocytoma Treatment for information about the treatment of osteosarcoma.) Ewing Sarcoma The detection of a translocation involving the EWSR1 gene on chromosome 22 band q12 and any one of a number of partner chromosomes is the key feature in the diagnosis of Ewing sarcoma (refer to Table 2).[17] The EWSR1 gene is a member of the TET family [TLS/EWS/TAF15] of RNA-binding proteins.[18] The FLI1 gene is a member of the ETS family of DNA-binding genes. Characteristically, the amino terminus of the EWSR1 gene is juxtaposed with the carboxy terminus of the STS family gene. In most cases (90%), the carboxy terminus is provided by FLI1, a member of the family of transcription factor genes located on chromosome 11 band q24. Other family members that may combine with the EWSR1 gene are ERG, ETV1, ETV4 (also termed E1AF), and FEV.[19] Rarely, TLS, another TET family member, can substitute for EWSR1.[20] Finally, there are a few rare cases in which EWSR1 has translocated with partners that are not members of the ETS family of oncogenes. The significance of these alternate partners is not known. Besides these consistent aberrations involving the EWSR1 gene at 22q12, additional numerical and structural aberrations have been observed in Ewing sarcoma, including gains of chromosomes 2, 5, 8, 9, 12, and 15; the nonreciprocal translocation t(1;16)(q12;q11.2); and deletions on the short arm of chromosome 6. Trisomy 20 may be associated with a more aggressive subset of Ewing sarcoma.[21] Three papers have described the genomic landscape of Ewing sarcoma and all show that these tumors have a relatively silent genome, with a paucity of mutations in pathways that might be amenable to treatment with novel targeted therapies.[22,23,24] These papers also identified mutations in STAG2, a member of the cohesin complex, in about 15% to 20% of the cases, and the presence of these mutations was associated with advanced-stage disease. CDKN2A deletions were noted in 12% to 22% of cases. Finally, TP53 mutations were identified in about 6% to 7% of cases and the coexistence of STAG2 and TP53 mutations is associated with a poor clinical outcome.[22,23,24] Figure 10 below from a discovery cohort (n = 99) highlights the frequency of chromosome 8 gain, the co-occurrence of chromosome 1q gain and chromosome 16q loss, the mutual exclusivity of CDKN2A deletion and STAG2 mutation, and the relative paucity of recurrent single nucleotide variants for Ewing sarcoma.[22]
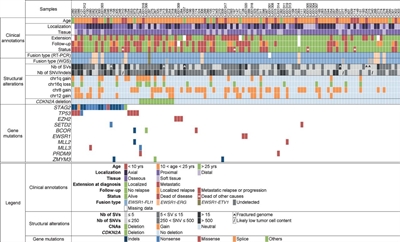
Figure 10. A comprehensive profile of the genetic abnormalities in Ewing sarcoma and associated clinical information. Key clinical characteristics are indicated, including primary site, type of tissue, and metastatic status at diagnosis, follow-up, and last news. Below is the consistency of detection of gene fusions by RT-PCR and whole-genome sequencing (WGS). The numbers of structural variants (SV) and single-nucleotide variants (SNV) as well as indels are reported in grayscale. The presence of the main copy-number changes, chr 1q gain, chr 16 loss, chr 8 gain, chr 12 gain, and interstitial CDKN2A deletion is indicated. Listed last are the most significant mutations and their types. For gene mutations, "others" refers to: duplication of exon 22 leading to frameshift (STAG2), deletion of exon 2 to 11 (BCOR), and deletion of exons 1 to 6 (ZMYM3). Reprinted from Cancer Discovery, Copyright 2014, 4 (11), 1342-53, Tirode F, Surdez D, Ma X, et al., Genomic Landscape of Ewing Sarcoma Defines an Aggressive Subtype with Co-Association of STAG2 and TP53 mutations, with permission from AACR.
Ewing sarcoma translocations can all be found with standard cytogenetic analysis. A more rapid analysis looking for a break apart of the EWS gene is now frequently done to confirm the diagnosis of Ewing sarcoma molecularly.[25] This test result must be considered with caution, however. Ewing sarcomas that utilize the TLS translocations will have negative tests because the EWSR1 gene is not translocated in those cases. In addition, other small round tumors also contain translocations of different ETS family members with EWSR1, such as desmoplastic small round cell tumor, clear cell sarcoma, extraskeletal myxoid chondrosarcoma, and myxoid liposarcoma, all of which may be positive with a EWS fluorescence in situ hybridization (FISH) break-apart probe. A detailed analysis of 85 patients with small round blue cell tumors that were negative for EWSR1 rearrangement by FISH with an EWSR1 break-apart probe identified eight patients with FUS rearrangements.[26] Four patients who had EWSR1-ERG fusions were not detected by FISH with an EWSR1 break-apart probe. The authors do not recommend relying solely on EWSR1 break-apart probes for analyzing small round blue cell tumors with strong immunohistochemical positivity for CD99. Small round blue cell tumors of bone and soft tissue that are histologically similar to Ewing sarcoma but do not have rearrangements of the EWSR1 gene have been analyzed and translocations have been identified. These include BCOR-CCNB3, CIC-DUX4, and CIC-FOX4.[27,28,29,30] The molecular profile of these tumors is different from the profile of EWS-FLI1 translocated Ewing sarcoma, and limited evidence suggests that they have a different clinical behavior. In almost all cases, the patients were treated with therapy designed for Ewing sarcoma on the basis of the histologic and immunohistologic similarity to Ewing sarcoma. There are too few cases associated with each translocation to determine whether the prognosis for these small round blue cell tumors is distinct from the prognosis of Ewing sarcoma of similar stage and site.[27,28,29,30] A genome-wide association study identified a region on chromosome 10q21.3 associated with an increased risk of Ewing sarcoma.[31] Deep sequencing through this region identified a polymorphism in the EGR2 gene, which appears to cooperate with the gene product of the EWSR1-FLI1 fusion that is seen in most patients with Ewing sarcoma.[32] The polymorphism associated with the increased risk is found at a much higher frequency in whites than in blacks or Asians, possibly contributing to the epidemiology of the relative infrequency of Ewing sarcoma in the latter populations. Table 2.EWSandTLSFusions and Translocations in Ewing Sarcoma| TET Family Partner | Fusion With ETS-like Oncogene Partner | Translocation | Comment |
|---|
| a These partners are not members of theETSfamily of oncogenes. | | EWS | EWSR1-FLI1 | t(11;22)(q24;q12) | Most common; ~85% to 90% of cases | | EWSR1-ERG | t(21;22)(q22;q12) | Second most common; ~10% of cases | | EWSR1-ETV1 | t(7;22)(p22;q12) | Rare | | EWSR1-ETV4 | t(17;22)(q12;q12) | Rare | | EWSR1-FEV | t(2;22)(q35;q12) | Rare | | EWSR1-NFATc2a | t(20;22)(q13;q12) | Rare | | EWSR1-POU5F1a | t(6;22)(p21;q12) | | | EWSR1-SMARCA5a | t(4;22)(q31;q12) | Rare | | EWSR1-ZSGa | t(6;22)(p21;q12) | | | EWSR1-SP3a | t(2;22)(q31;q12) | Rare | | TLS(also calledFUS) | TLS-ERG | t(16;21)(p11;q22) | Rare | | TLS-FEV | t(2;16)(q35;p11) | Rare | (Refer to the PDQ summary on Ewing Sarcoma Treatment for information about the treatment of Ewing sarcoma.) Rhabdomyosarcoma The embryonal and alveolar histologies have distinctive molecular characteristics that have been used for diagnostic confirmation, and may be useful for assigning risk group, determining therapy, and monitoring residual disease during treatment.[33,34,35,36,37] - Embryonal histology: Embryonal tumors often show loss of heterozygosity at 11p15 and gains on chromosome 8.[38,39,40] Embryonal tumors have a higher background mutation rate and higher single-nucleotide variant rate than do alveolar tumors, and the number of somatic mutations increases with older age at diagnosis.[41,42] Genes with recurring mutations include those in the RAS pathway (e.g., NRAS, KRAS, HRAS, and NF1), which together are observed in approximately one-third of cases. Other genes with recurring mutations include FGFR4, PIK3CA, CTNNB1, FBXW7, and BCOR, all of which are present in fewer than 10% of cases.[41,42]
Embryonal histology with anaplasia: Anaplasia has been reported in a minority of children with rhabdomyosarcoma, primarily arising in children with the embryonal subtype who are younger than 10 years.[43,44] Rhabdomyosarcoma with nonalveolar, anaplastic morphology may be a presenting feature for children with Li-Fraumeni syndrome and germline TP53 mutations.[45] Among eight consecutively presenting children with rhabdomyosarcoma and TP53 germline mutations, all showed anaplastic morphology. Among an additional seven children with anaplastic rhabdomyosarcoma and unknown TP53 germline mutation status, three of the seven children had functionally relevant TP53 germline mutations. The median age at diagnosis of the 11 children with TP53 germline mutation status was 40 months (range, 19-67 months). - Alveolar histology: About 70% to 80% of alveolar tumors are characterized by translocations between the FOXO1 gene on chromosome 13 and either the PAX3 gene on chromosome 2 (t(2;13)(q35;q14)) or the PAX7 gene on chromosome 1 (t(1;13)(p36;q14)).[33,38,46] Other rare fusions include PAX3-NCOA1 and PAX3-INO80D.[41] Translocations involving the PAX3 gene occur in approximately 59% of alveolar rhabdomyosarcoma cases, while the PAX7 gene appears to be involved in about 19% of cases.[33] Patients with solid-variant alveolar histology have a lower incidence of PAX-FOXO1 gene fusions than do patients showing classical alveolar histology.[47] For the diagnosis of alveolar rhabdomyosarcoma, FOXO1 gene rearrangement may be detected with good sensitivity and specificity using either fluorescence in situ hybridization or reverse transcription-polymerase chain reaction.[48]
The alveolar histology that is associated with the PAX7 gene in patients with or without metastatic disease appears to occur at a younger age and may be associated with longer event-free survival rates than those associated with PAX3 gene rearrangements.[49,50,51,52,53,54] Patients with alveolar histology and the PAX3 gene are older and have a higher incidence of invasive tumor (T2). Around 22% of cases showing alveolar histology have no detectable PAX gene translocation.[37,47] In addition to FOXO1 rearrangements, alveolar tumors are characterized by a lower mutational burden than are fusion-negative tumors, with fewer genes having recurring mutations.[41,42]BCOR and PIK3CA mutations and amplification of MYCN, MIR17HG, and CDK4 have also been described. - Spindle cell/sclerosing histology: Spindle cell/sclerosing rhabdomyosarcoma has been proposed as a separate entity in the World Health Organization Classification of Tumours of Soft Tissue and Bone.[55] For congenital/infantile spindle cell rhabdomyosarcoma, a study reported that 10 of 11 patients showed recurrent fusion genes. Most of these cases had truncal primary tumors, and no paratesticular tumors were found. Novel VGLL2 rearrangements were observed in seven patients (63%), including VGLL2-CITED2 fusion in four patients and VGLL2-NCOA2 in two patients.[56] Three patients (27%) harbored different NCOA2 gene fusions, including TEAD1-NCOA2 in two patients and SRF-NCOA2 in one patient. All fusion-positive congenital/infantile spindle cell rhabdomyosarcoma patients with available long-term follow-up were alive and well, and no patients developed distant metastases.[56] Further study is needed to better define the prevalence and prognostic significance of these gene rearrangements in young children with spindle cell rhabdomyosarcoma.
In older children and adults with spindle cell/sclerosing rhabdomyosarcoma, a specific MYOD1 mutation (p.L122R) has been observed in a large proportion of patients.[56,57,58,59] Activating PIK3CA mutations were common in MYOD1-mutated cases (4 of 10); when they were present, they were associated with sclerosing histology.[56] The presence of the MYOD1 mutation is associated with an increased risk of treatment failure.[56,57,58] In one study that included nine children aged 1 year or older with spindle cell/sclerosing histology and MYOD1 mutations, seven had a fatal outcome despite aggressive multimodality treatment.[56]
These findings highlight the important differences between embryonal and alveolar tumors. Data demonstrate that PAX-FOX01 fusion-positive alveolar tumors are biologically and clinically different from fusion-negative alveolar tumors and embryonal tumors.[37,60,61,62,63] In a study of Intergroup Rhabdomyosarcoma Study Group cases, which captured an entire cohort from a single prospective clinical trial, the outcome for patients with translocation-negative alveolar rhabdomyosarcoma was better than that observed for translocation-positive cases. The outcome was similar to that seen in patients with embryonal rhabdomyosarcoma and demonstrated that fusion status is a critical factor for risk stratification in pediatric rhabdomyosarcoma. (Refer to the PDQ summary on Childhood Rhabdomyosarcoma Treatment for information about the treatment of childhood rhabdomyosarcoma.) References:
-
Chen X, Bahrami A, Pappo A, et al.: Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep 7 (1): 104-12, 2014.
-
Perry JA, Kiezun A, Tonzi P, et al.: Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A 111 (51): E5564-73, 2014.
-
Ognjanovic S, Olivier M, Bergemann TL, et al.: Sarcomas in TP53 germline mutation carriers: a review of the IARC TP53 database. Cancer 118 (5): 1387-96, 2012.
-
Mirabello L, Yeager M, Mai PL, et al.: Germline TP53 variants and susceptibility to osteosarcoma. J Natl Cancer Inst 107 (7): , 2015.
-
Toguchida J, Yamaguchi T, Dayton SH, et al.: Prevalence and spectrum of germline mutations of the p53 gene among patients with sarcoma. N Engl J Med 326 (20): 1301-8, 1992.
-
McIntyre JF, Smith-Sorensen B, Friend SH, et al.: Germline mutations of the p53 tumor suppressor gene in children with osteosarcoma. J Clin Oncol 12 (5): 925-30, 1994.
-
Kansara M, Thomas DM: Molecular pathogenesis of osteosarcoma. DNA Cell Biol 26 (1): 1-18, 2007.
-
German J: Bloom's syndrome. XX. The first 100 cancers. Cancer Genet Cytogenet 93 (1): 100-6, 1997.
-
Lipton JM, Federman N, Khabbaze Y, et al.: Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol 23 (1): 39-44, 2001.
-
Idol RA, Robledo S, Du HY, et al.: Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis 39 (1): 35-43, 2007 Jul-Aug.
-
Li FP, Fraumeni JF Jr, Mulvihill JJ, et al.: A cancer family syndrome in twenty-four kindreds. Cancer Res 48 (18): 5358-62, 1988.
-
Grimer RJ, Cannon SR, Taminiau AM, et al.: Osteosarcoma over the age of forty. Eur J Cancer 39 (2): 157-63, 2003.
-
Wong FL, Boice JD Jr, Abramson DH, et al.: Cancer incidence after retinoblastoma. Radiation dose and sarcoma risk. JAMA 278 (15): 1262-7, 1997.
-
Wang LL, Gannavarapu A, Kozinetz CA, et al.: Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst 95 (9): 669-74, 2003.
-
Hicks MJ, Roth JR, Kozinetz CA, et al.: Clinicopathologic features of osteosarcoma in patients with Rothmund-Thomson syndrome. J Clin Oncol 25 (4): 370-5, 2007.
-
Goto M, Miller RW, Ishikawa Y, et al.: Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol Biomarkers Prev 5 (4): 239-46, 1996.
-
Delattre O, Zucman J, Melot T, et al.: The Ewing family of tumors--a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med 331 (5): 294-9, 1994.
-
Urano F, Umezawa A, Yabe H, et al.: Molecular analysis of Ewing's sarcoma: another fusion gene, EWS-E1AF, available for diagnosis. Jpn J Cancer Res 89 (7): 703-11, 1998.
-
Hattinger CM, Rumpler S, Strehl S, et al.: Prognostic impact of deletions at 1p36 and numerical aberrations in Ewing tumors. Genes Chromosomes Cancer 24 (3): 243-54, 1999.
-
Sankar S, Lessnick SL: Promiscuous partnerships in Ewing's sarcoma. Cancer Genet 204 (7): 351-65, 2011.
-
Roberts P, Burchill SA, Brownhill S, et al.: Ploidy and karyotype complexity are powerful prognostic indicators in the Ewing's sarcoma family of tumors: a study by the United Kingdom Cancer Cytogenetics and the Children's Cancer and Leukaemia Group. Genes Chromosomes Cancer 47 (3): 207-20, 2008.
-
Tirode F, Surdez D, Ma X, et al.: Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 4 (11): 1342-53, 2014.
-
Crompton BD, Stewart C, Taylor-Weiner A, et al.: The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 4 (11): 1326-41, 2014.
-
Brohl AS, Solomon DA, Chang W, et al.: The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 10 (7): e1004475, 2014.
-
Monforte-Muñoz H, Lopez-Terrada D, Affendie H, et al.: Documentation of EWS gene rearrangements by fluorescence in-situ hybridization (FISH) in frozen sections of Ewing's sarcoma-peripheral primitive neuroectodermal tumor. Am J Surg Pathol 23 (3): 309-15, 1999.
-
Chen S, Deniz K, Sung YS, et al.: Ewing sarcoma with ERG gene rearrangements: A molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH. Genes Chromosomes Cancer 55 (4): 340-9, 2016.
-
Pierron G, Tirode F, Lucchesi C, et al.: A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet 44 (4): 461-6, 2012.
-
Specht K, Sung YS, Zhang L, et al.: Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: further evidence toward distinct pathologic entities. Genes Chromosomes Cancer 53 (7): 622-33, 2014.
-
Sugita S, Arai Y, Tonooka A, et al.: A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol 38 (11): 1571-6, 2014.
-
Cohen-Gogo S, Cellier C, Coindre JM, et al.: Ewing-like sarcomas with BCOR-CCNB3 fusion transcript: a clinical, radiological and pathological retrospective study from the Société Française des Cancers de L'Enfant. Pediatr Blood Cancer 61 (12): 2191-8, 2014.
-
Postel-Vinay S, Véron AS, Tirode F, et al.: Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet 44 (3): 323-7, 2012.
-
Grünewald TG, Bernard V, Gilardi-Hebenstreit P, et al.: Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet 47 (9): 1073-8, 2015.
-
Barr FG, Smith LM, Lynch JC, et al.: Examination of gene fusion status in archival samples of alveolar rhabdomyosarcoma entered on the Intergroup Rhabdomyosarcoma Study-III trial: a report from the Children's Oncology Group. J Mol Diagn 8 (2): 202-8, 2006.
-
Kelly KM, Womer RB, Barr FG: Minimal disease detection in patients with alveolar rhabdomyosarcoma using a reverse transcriptase-polymerase chain reaction method. Cancer 78 (6): 1320-7, 1996.
-
Edwards RH, Chatten J, Xiong QB, et al.: Detection of gene fusions in rhabdomyosarcoma by reverse transcriptase-polymerase chain reaction assay of archival samples. Diagn Mol Pathol 6 (2): 91-7, 1997.
-
Sartori F, Alaggio R, Zanazzo G, et al.: Results of a prospective minimal disseminated disease study in human rhabdomyosarcoma using three different molecular markers. Cancer 106 (8): 1766-75, 2006.
-
Davicioni E, Anderson MJ, Finckenstein FG, et al.: Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group. Am J Pathol 174 (2): 550-64, 2009.
-
Merlino G, Helman LJ: Rhabdomyosarcoma--working out the pathways. Oncogene 18 (38): 5340-8, 1999.
-
Koufos A, Hansen MF, Copeland NG, et al.: Loss of heterozygosity in three embryonal tumours suggests a common pathogenetic mechanism. Nature 316 (6026): 330-4, 1985 Jul 25-31.
-
Scrable H, Witte D, Shimada H, et al.: Molecular differential pathology of rhabdomyosarcoma. Genes Chromosomes Cancer 1 (1): 23-35, 1989.
-
Shern JF, Chen L, Chmielecki J, et al.: Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 4 (2): 216-31, 2014.
-
Chen X, Stewart E, Shelat AA, et al.: Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell 24 (6): 710-24, 2013.
-
Kodet R, Newton WA Jr, Hamoudi AB, et al.: Childhood rhabdomyosarcoma with anaplastic (pleomorphic) features. A report of the Intergroup Rhabdomyosarcoma Study. Am J Surg Pathol 17 (5): 443-53, 1993.
-
Qualman S, Lynch J, Bridge J, et al.: Prevalence and clinical impact of anaplasia in childhood rhabdomyosarcoma : a report from the Soft Tissue Sarcoma Committee of the Children's Oncology Group. Cancer 113 (11): 3242-7, 2008.
-
Hettmer S, Archer NM, Somers GR, et al.: Anaplastic rhabdomyosarcoma in TP53 germline mutation carriers. Cancer 120 (7): 1068-75, 2014.
-
Dumont SN, Lazar AJ, Bridge JA, et al.: PAX3/7-FOXO1 fusion status in older rhabdomyosarcoma patient population by fluorescent in situ hybridization. J Cancer Res Clin Oncol 138 (2): 213-20, 2012.
-
Parham DM, Qualman SJ, Teot L, et al.: Correlation between histology and PAX/FKHR fusion status in alveolar rhabdomyosarcoma: a report from the Children's Oncology Group. Am J Surg Pathol 31 (6): 895-901, 2007.
-
Thway K, Wang J, Wren D, et al.: The comparative utility of fluorescence in situ hybridization and reverse transcription-polymerase chain reaction in the diagnosis of alveolar rhabdomyosarcoma. Virchows Arch 467 (2): 217-24, 2015.
-
Sorensen PH, Lynch JC, Qualman SJ, et al.: PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol 20 (11): 2672-9, 2002.
-
Krsková L, Mrhalová M, Sumerauer D, et al.: Rhabdomyosarcoma: molecular diagnostics of patients classified by morphology and immunohistochemistry with emphasis on bone marrow and purged peripheral blood progenitor cells involvement. Virchows Arch 448 (4): 449-58, 2006.
-
Kelly KM, Womer RB, Sorensen PH, et al.: Common and variant gene fusions predict distinct clinical phenotypes in rhabdomyosarcoma. J Clin Oncol 15 (5): 1831-6, 1997.
-
Barr FG, Qualman SJ, Macris MH, et al.: Genetic heterogeneity in the alveolar rhabdomyosarcoma subset without typical gene fusions. Cancer Res 62 (16): 4704-10, 2002.
-
Missiaglia E, Williamson D, Chisholm J, et al.: PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol 30 (14): 1670-7, 2012.
-
Duan F, Smith LM, Gustafson DM, et al.: Genomic and clinical analysis of fusion gene amplification in rhabdomyosarcoma: a report from the Children's Oncology Group. Genes Chromosomes Cancer 51 (7): 662-74, 2012.
-
Nascimento AF, Barr FG, Fletcher CD, et al., eds.: Spindle cell/sclerosing rhabdomyosarcoma. In: Fletcher CDM, Bridge JA, Hogendoorn P, et al., eds.: WHO Classification of Tumours of Soft Tissue and Bone. 4th ed. Lyon, France: IARC Press, 2013, pp 134-5.
-
Alaggio R, Zhang L, Sung YS, et al.: A Molecular Study of Pediatric Spindle and Sclerosing Rhabdomyosarcoma: Identification of Novel and Recurrent VGLL2-related Fusions in Infantile Cases. Am J Surg Pathol 40 (2): 224-35, 2016.
-
Kohsaka S, Shukla N, Ameur N, et al.: A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet 46 (6): 595-600, 2014.
-
Agaram NP, Chen CL, Zhang L, et al.: Recurrent MYOD1 mutations in pediatric and adult sclerosing and spindle cell rhabdomyosarcomas: evidence for a common pathogenesis. Genes Chromosomes Cancer 53 (9): 779-87, 2014.
-
Szuhai K, de Jong D, Leung WY, et al.: Transactivating mutation of the MYOD1 gene is a frequent event in adult spindle cell rhabdomyosarcoma. J Pathol 232 (3): 300-7, 2014.
-
Davicioni E, Anderson JR, Buckley JD, et al.: Gene expression profiling for survival prediction in pediatric rhabdomyosarcomas: a report from the children's oncology group. J Clin Oncol 28 (7): 1240-6, 2010.
-
Williamson D, Missiaglia E, de Reyniès A, et al.: Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol 28 (13): 2151-8, 2010.
-
Davicioni E, Finckenstein FG, Shahbazian V, et al.: Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res 66 (14): 6936-46, 2006.
-
Skapek SX, Anderson J, Barr FG, et al.: PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children's oncology group report. Pediatr Blood Cancer 60 (9): 1411-7, 2013.
Langerhans Cell HistiocytosisStudies published in 1994 showed clonality in Langerhans cell histiocytosis (LCH) using polymorphisms of methylation-specific restriction enzyme sites on the X-chromosome regions coding for the human androgen receptor, DXS255, PGK, and HPRT.[1,2] Biopsies of lesions with single-system or multisystem disease were found to have a proliferation of LCH cells from a single clone. The discovery of recurring genomic alterations (primarily BRAF V600E) in LCH (see below) confirmed the clonality of LCH in children. Pulmonary LCH in adults is usually nonclonal and it is possible that this group represents a reactive process to smoking.[3] However, a subset appeared to be clonal, as an analysis of BRAF mutations showed that a significant proportion of patients (25%-30%) have evidence for mutant BRAF V600E.[4]
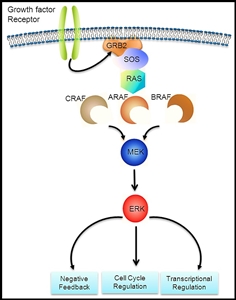
Figure 11. Courtesy of Rikhia Chakraborty, Ph.D. Permission to reuse the figure in any form must be obtained directly from Dr. Chakraborty.
The genomic basis of LCH was advanced by a report in 2010 of an activating mutation of the BRAF oncogene (V600E) that was detected in 35 (57%) of 61 cases.[5] Multiple subsequent reports have confirmed the presence of BRAF V600E mutations in 50% or more of LCH cases in children.[6,7,8] Another BRAF mutation (BRAF 600DLAT) was identified, which resulted in the insertion of four amino acids and also appeared to activate signaling.[7]ARAF mutations are infrequent in LCH, but when present, can also lead to RAS-MAPK pathway activation.[9] No clinical characteristics associated with the BRAF V600E mutation have been identified.[5,6,7] The RAS-MAPK signaling pathway (Figure 11) transmits signals from a cell surface receptor (e.g., a growth factor) through the RAS pathway (via one of the RAF proteins [A, B, or C]) to phosphorylate MEK and then the extracellular signal-regulated kinase (ERK), which leads to nuclear signals affecting cell cycle and transcription regulation. The V600E mutation of BRAF leads to continuous phosphorylation, and thus activation, of MEK and ERK without the need for an external signal. Activation of ERK occurs by phosphorylation, and phosphorylated ERK can be detected in virtually all LCH lesions.[5,10] Because RAS-MAPK pathway activation can be detected in all LCH cases, but not all cases have BRAF mutations, the presence of genomic alterations in other components of the pathway was suspected. Whole-exome sequencing of BRAF-mutated versus BRAF-wild-type LCH biopsies revealed that 7 of 21 BRAF-wild-type specimens had MAP2K1 mutations, while no BRAF-mutated specimens had MAP2K1 mutations.[10] The mutations in MAP2K1 (which codes for MEK) were activating, as indicated by their induction of ERK phosphorylation.[10] Another study showed MAP2K1 mutations exclusively in 11 of 22 BRAF-wild-type cases.[11] Finally, in-frame BRAF deletions and in-frame FAM73A-BRAF fusions have occurred in the group of BRAF V600E and MAP2K1 mutation-negative cases.[12] Studies to date support the universal activation of ERK in LCH, with activation in most cases being explained by BRAF and MAP2K1 alterations.[5,10,12] The presence of BRAF V600E mutation in blood and bone marrow was studied in a series of 100 patients, of which 65% tested positive for the BRAF V600E mutation by a sensitive quantitative polymerase chain reaction technique.[6] Circulating cells with the BRAF V600E mutation could be detected in all high-risk patients and in a subset of low-risk multisystem patients. The presence of circulating cells with the mutation conferred a twofold increased risk of relapse. The myeloid dendritic cell origin of LCH was confirmed by finding CD34+ stem cells with the mutation in the bone marrow of high-risk patients. Those with low-risk disease had more mature myeloid dendritic cells with the mutation, suggesting the stage of cell development is critical in defining the clinical characteristics of LCH, which can now be considered a myeloid neoplasia in most cases. A study of 173 patients with the BRAF V600E mutation, and 142 patients without the mutation, revealed that the mutation occurred in 88% of patients with high-risk disease, 69% of patients with multisystem low-risk LCH, and 44% of patients with single-system low-risk LCH.[13] The mutation was also found in 75% of patients with neurodegenerative syndrome and 73% of patients with pituitary involvement. Resistance to initial treatment and relapse were higher in patients with the mutation.[13] Clinical implications Clinical implications of the described genomic findings include the following: - LCH joins a group of other pediatric entities with activating BRAF mutations, including select nonmalignant conditions (e.g., benign nevi) [14] and low-grade malignancies (e.g., pilocytic astrocytoma).[15,16] All of these conditions have a generally indolent course, with spontaneous resolution occurring in some cases. This distinctive clinical course may be a manifestation of oncogene-induced senescence.[14,17]
- BRAF V600E mutations can be targeted by BRAF inhibitors (e.g., vemurafenib and dabrafenib) or by the combination of BRAF inhibitors plus MEK inhibitors (e.g., dabrafenib/trametinib and vemurafenib/cobimetinib). These agents and combinations are approved for adults with melanoma. Treatment of adults with combinations of a BRAF inhibitor and a MEK inhibitor showed significantly improved progression-free survival outcome compared with treatment using a BRAF inhibitor alone.[18,19] The most serious side effect of BRAF inhibitors is the induction of cutaneous squamous cell carcinomas,[18,19] with the incidence of these second cancers increasing with age;[20] reduction of this side effect can occur with concurrent treatment with both BRAF and MEK inhibitors.[18,19] Case reports have described activity of BRAF inhibitors against LCH in adult [21,22,23,24,25] and pediatric [26] patients, but there are insufficient data to assess the role of these agents in treatment of children with LCH.
- With further research, the observation of BRAF V600E (or potentially mutated MAP2K1) in circulating cells may become a useful diagnostic tool to define high-risk versus low-risk disease.[6] Additionally, for patients who have a somatic mutation, persistence of circulating cells with the mutation may be useful as a marker of residual disease.[6]
(Refer to the PDQ summary on Langerhans Cell Histiocytosis Treatment for information about the treatment of childhood LCH.) References:
-
Willman CL, Busque L, Griffith BB, et al.: Langerhans'-cell histiocytosis (histiocytosis X)--a clonal proliferative disease. N Engl J Med 331 (3): 154-60, 1994.
-
Yu RC, Chu C, Buluwela L, et al.: Clonal proliferation of Langerhans cells in Langerhans cell histiocytosis. Lancet 343 (8900): 767-8, 1994.
-
Dacic S, Trusky C, Bakker A, et al.: Genotypic analysis of pulmonary Langerhans cell histiocytosis. Hum Pathol 34 (12): 1345-9, 2003.
-
Roden AC, Hu X, Kip S, et al.: BRAF V600E expression in Langerhans cell histiocytosis: clinical and immunohistochemical study on 25 pulmonary and 54 extrapulmonary cases. Am J Surg Pathol 38 (4): 548-51, 2014.
-
Badalian-Very G, Vergilio JA, Degar BA, et al.: Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood 116 (11): 1919-23, 2010.
-
Berres ML, Lim KP, Peters T, et al.: BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J Exp Med 211 (4): 669-83, 2014.
-
Satoh T, Smith A, Sarde A, et al.: B-RAF mutant alleles associated with Langerhans cell histiocytosis, a granulomatous pediatric disease. PLoS One 7 (4): e33891, 2012.
-
Sahm F, Capper D, Preusser M, et al.: BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood 120 (12): e28-34, 2012.
-
Nelson DS, Quispel W, Badalian-Very G, et al.: Somatic activating ARAF mutations in Langerhans cell histiocytosis. Blood 123 (20): 3152-5, 2014.
-
Chakraborty R, Hampton OA, Shen X, et al.: Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood 124 (19): 3007-15, 2014.
-
Brown NA, Furtado LV, Betz BL, et al.: High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative Langerhans cell histiocytosis. Blood 124 (10): 1655-8, 2014.
-
Chakraborty R, Burke TM, Hampton OA, et al.: Alternative genetic mechanisms of BRAF activation in Langerhans cell histiocytosis. Blood 128 (21): 2533-2537, 2016.
-
Héritier S, Emile JF, Barkaoui MA, et al.: BRAF Mutation Correlates With High-Risk Langerhans Cell Histiocytosis and Increased Resistance to First-Line Therapy. J Clin Oncol 34 (25): 3023-30, 2016.
-
Michaloglou C, Vredeveld LC, Soengas MS, et al.: BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436 (7051): 720-4, 2005.
-
Jones DT, Kocialkowski S, Liu L, et al.: Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68 (21): 8673-7, 2008.
-
Pfister S, Janzarik WG, Remke M, et al.: BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest 118 (5): 1739-49, 2008.
-
Jacob K, Quang-Khuong DA, Jones DT, et al.: Genetic aberrations leading to MAPK pathway activation mediate oncogene-induced senescence in sporadic pilocytic astrocytomas. Clin Cancer Res 17 (14): 4650-60, 2011.
-
Larkin J, Ascierto PA, Dréno B, et al.: Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 371 (20): 1867-76, 2014.
-
Long GV, Stroyakovskiy D, Gogas H, et al.: Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet 386 (9992): 444-51, 2015.
-
Anforth R, Menzies A, Byth K, et al.: Factors influencing the development of cutaneous squamous cell carcinoma in patients on BRAF inhibitor therapy. J Am Acad Dermatol 72 (5): 809-15.e1, 2015.
-
Haroche J, Cohen-Aubart F, Emile JF, et al.: Reproducible and sustained efficacy of targeted therapy with vemurafenib in patients with BRAF(V600E)-mutated Erdheim-Chester disease. J Clin Oncol 33 (5): 411-8, 2015.
-
Charles J, Beani JC, Fiandrino G, et al.: Major response to vemurafenib in patient with severe cutaneous Langerhans cell histiocytosis harboring BRAF V600E mutation. J Am Acad Dermatol 71 (3): e97-9, 2014.
-
Gandolfi L, Adamo S, Pileri A, et al.: Multisystemic and Multiresistant Langerhans Cell Histiocytosis: A Case Treated With BRAF Inhibitor. J Natl Compr Canc Netw 13 (6): 715-8, 2015.
-
Euskirchen P, Haroche J, Emile JF, et al.: Complete remission of critical neurohistiocytosis by vemurafenib. Neurol Neuroimmunol Neuroinflamm 2 (2): e78, 2015.
-
Hyman DM, Puzanov I, Subbiah V, et al.: Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 373 (8): 726-36, 2015.
-
Héritier S, Jehanne M, Leverger G, et al.: Vemurafenib Use in an Infant for High-Risk Langerhans Cell Histiocytosis. JAMA Oncol 1 (6): 836-8, 2015.
NeuroblastomaNeuroblastoma can be subdivided into a biologically defined subset that has a very favorable prognosis (i.e., low-risk neuroblastoma) and another group that has a guarded prognosis (i.e., high-risk neuroblastoma). While neuroblastoma in infants with tumors that have favorable biology is highly curable, only 50% of children with high-risk neuroblastoma are alive at 5 years from diagnosis, at best. Low-risk neuroblastoma is usually found in children younger than 18 months with limited extent of disease; the tumor has changes, usually increases, in the number of whole chromosomes in the neuroblastoma cell. Low-risk tumors are hyperdiploid when examined by flow cytometry.[1,2] In contrast, high-risk neuroblastoma generally occurs in children older than 18 months, is often metastatic to bone, and usually has segmental chromosome abnormalities. They are near diploid or near tetraploid by flow cytometric measurement.[1,2,3,4,5,6,7] High-risk tumors also show exonic mutations (refer to the Exonic mutations in neuroblastoma section of this summary for more information), but most high-risk tumors lack mutations in genes that are recurrently mutated. Compared with adult cancers, neuroblastomas show a low number of mutations per genome that affect protein sequence (10-20 per genome).[8] Key genomic characteristics of high-risk neuroblastoma that are discussed below include the following: - Segmental chromosomal aberrations, including MYCN gene amplification.
- Low rates of exonic mutations, with activating mutations in ALK being the most common recurring alteration.
- Genomic alterations that promote telomere lengthening.
Segmental chromosomal aberrations (includingMYCNgene amplification) Segmental chromosomal aberrations, found most frequently in 1p, 1q, 3p, 11q, 14q, and 17p (and MYCN amplification), are best detected by comparative genomic hybridization and are seen in almost all high-risk and/or stage 4 neuroblastomas.[3,4,5,6,7] Among all patients with neuroblastoma, a higher number of chromosome breakpoints correlated with the following, whether or not MYCN amplification was considered:[3,4,5,6,7][Level of evidence: 3iiD] - Advanced age at diagnosis.
- Advanced stage of disease.
- Higher risk of relapse.
- Poorer outcome.
In a study of unresectable primary neuroblastomas without metastases in children older than 12 months, segmental chromosomal aberrations were found in most, and older children were more likely to have them and to have more of them per tumor cell. In children aged 12 to 18 months, the presence of segmental chromosomal aberrations had a significant effect on event-free survival (EFS) but not on overall survival (OS). However, in children older than 18 months, there was a significant difference in OS in children with segmental chromosomal aberrations versus children without segmental chromosomal aberrations (67% vs. 100%), regardless of the histologic prognosis.[7] Segmental chromosomal aberrations are also predictive of recurrence in infants with localized unresectable or metastatic neuroblastoma without MYCN gene amplification.[1,2] MYCN amplification (defined as more than 10 copies per diploid genome) is one of the most common segmental chromosomal aberrations, detected in 16% to 25% of tumors.[9] For high-risk neuroblastoma, 40% to 50% of cases show MYCN amplification.[10] In all stages of disease, amplification of the MYCN gene strongly predicts a poorer prognosis in both time to tumor progression and OS in almost all multivariate regression analyses of prognostic factors.[1,2] Within the localized MYCN-amplified cohort, ploidy status may further predict outcome.[11] However, patients with hyperdiploid tumors with any segmental chromosomal aberrations do relatively poorly.[3] Most unfavorable clinical and pathobiological features are associated, to some degree, with MYCN amplification; in a multivariable logistic regression analysis of 7,102 International Neuroblastoma Risk Group patients, pooled segmental chromosomal aberrations and gain of 17q were the only poor prognostic features not associated with MYCN amplification. However, segmental chromosomal aberrations at 11q are almost mutually exclusive of MYCN amplification. Exonic mutations in neuroblastoma Multiple reports have documented that a minority of high-risk neuroblastomas have a small number of low-incidence, recurrently mutated genes. The most commonly mutated gene is ALK, which is mutated in approximately 10% of patients (see below). Other genes with even lower frequencies of mutation include ATRX, PTPN11, ARID1A, and ARID1B.[12,13,14,15,16,17,18] As shown in Figure 12, most neuroblastoma cases lack mutations in genes that are altered in a recurrent manner.
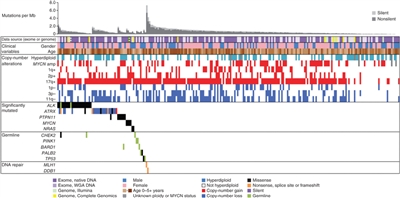
Figure 12. Data tracks (rows) facilitate the comparison of clinical and genomic data across cases with neuroblastoma (columns). The data sources and sequencing technology used were whole-exome sequencing (WES) from whole-genome amplification (WGA) (light purple), WES from native DNA (dark purple), Illumina WGS (green), and Complete Genomics WGS (yellow). Striped blocks indicate cases analyzed using two approaches. The clinical variables included were gender (male, blue; female, pink) and age (brown spectrum). Copy number alterations indicates ploidy measured by flow cytometry (with hyperdiploid meaning DNA index >1) and clinically relevant copy number alterations derived from sequence data. Significantly mutated genes are those with statistically significant mutation counts given the background mutation rate, gene size, and expression in neuroblastoma. Germline indicates genes with significant numbers of germline ClinVar variants or loss-of-function cancer gene variants in our cohort. DNA repair indicates genes that may be associated with an increased mutation frequency in two apparently hypermutated tumors. Predicted effects of somatic mutations are color coded according to the legend. Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics (Pugh TJ, Morozova O, Attiyeh EF, et al.: The genetic landscape of high-risk neuroblastoma. Nat Genet 45 (3): 279-84, 2013), copyright (2013).
ALK, the exonic mutation found most commonly in neuroblastoma, is a cell surface receptor tyrosine kinase, expressed at significant levels only in developing embryonic and neonatal brains. Germline mutations in ALK have been identified as the major cause of hereditary neuroblastoma. Somatically acquired ALK-activating mutations are also found as oncogenic drivers in neuroblastoma.[17] The presence of an ALK mutation correlates with significantly poorer survival in high-risk and intermediate-risk neuroblastoma patients. ALK mutation was examined in 1,596 diagnostic neuroblastoma samples.[17]ALK tyrosine kinase domain mutations occurred in 8% of samples-at three hot spots and 13 minor sites-and correlated significantly with poorer survival in patients with high-risk and intermediate-risk neuroblastoma. ALK mutations were found in 10.9% of MYCN-amplified tumors versus 7.2% of those without MYCN amplification. ALK mutations occurred at the highest frequency (11%) in patients older than 10 years.[17] The frequency of ALK aberrations was 14% in the high-risk neuroblastoma group, 6% in the intermediate-risk neuroblastoma group, and 8% in the low-risk neuroblastoma group. Small-molecule ALK kinase inhibitors such as crizotinib are being developed and tested in patients with recurrent and refractory neuroblastoma.[17] (Refer to the Treatment Options Under Clinical Evaluation for Recurrent or Refractory Neuroblastoma section in the PDQ summary on Neuroblastoma Treatment for more information about crizotinib clinical trials.) Genomic evolution of exonic mutations There are limited data regarding the genomic evolution of exonic mutations from diagnosis to relapse for neuroblastoma. Whole-genome sequencing was applied to 23 paired diagnostic and relapsed neuroblastomas to define somatic genetic alterations associated with relapse,[19] while a second study evaluated 16 paired diagnostic and relapsed specimens.[20] Both studies identified an increased number of mutations in the relapsed samples compared with the samples at diagnosis. - The first study found increased incidence of mutations in genes associated with RAS-MAPK signaling at relapse than at diagnosis, with 15 of 23 relapse samples containing somatic mutations in genes involved in this pathway and each mutation consistent with pathway activation.[19]
In addition, three relapse samples showed structural alterations involving MAPK pathway genes consistent with pathway activation, so aberrations in this pathway were detected in 18 of 23 relapse samples (78%). Aberrations were found in ALK (n = 10), NF1 (n = 2), and one each in NRAS, KRAS, HRAS, BRAF, PTPN11, and FGFR1. Even with deep sequencing, 7 of the 18 alterations were not detectable in the primary tumor, highlighting the evolution of mutation presumably leading to relapse and the importance of genomic evaluations of tissues obtained at relapse. - In the second study, ALK mutations were not observed in either diagnostic or relapse specimens, but relapse-specific recurrent single-nucleotide variants were observed in 11 genes, including the putative CHD5 neuroblastoma tumor suppressor gene located at chromosome 1p36.[20]
In a study of 276 neuroblastoma samples of all stages and from patients of all ages, very deep (33,000X) sequencing of two amplified ALK mutational hot spots revealed 4.8% clonal mutations and an additional 5% subclonal mutations, suggesting that subclonal mutations are common.[21] Deep sequencing can reveal the presence of mutations in tiny subsets of tumor cells that may be able to survive during treatment and grow to constitute a relapse. Genomic alterations promoting telomere lengthening Lengthening of telomeres, the tips of chromosomes, promotes cell survival. Telomeres otherwise shorten with each cell replication, resulting eventually in the lack of a cell's ability to replicate. Low-risk neuroblastomas have little telomere lengthening activity. Aberrant genetic mechanisms for telomere lengthening have been identified for high-risk neuroblastoma.[12,13,22] Thus far, the following three mechanisms, which appear to be mutually exclusive, have been described: - Chromosomal rearrangements involving a chromosomal region at 5p15.33 proximal to the TERT gene, which encodes the catalytic unit of telomerase, occur in approximately 25% of high-risk neuroblastoma cases and are mutually exclusive with MYCN amplifications and ATRX mutations.[12,13] The rearrangements induce transcriptional upregulation of TERT by juxtaposing the TERT coding sequence with strong enhancer elements.
- Another mechanism promoting TERT overexpression is MYCN amplification,[23] which is associated with approximately 40% to 50% of high-risk neuroblastomas.
- The ATRX mutation or deletion is found in 10% to 20% of high-risk neuroblastomas, almost exclusively in older children,[14] and is associated with telomere lengthening by a different mechanism, termed alternative lengthening of telomeres.[14,22]
Additional biological factors associated with prognosis MYC and MYCN expression Immunostaining for MYC and MYCN proteins on 357 undifferentiated/poorly differentiated neuroblastomas has demonstrated that elevated MYC/MYCN protein expression is prognostically significant.[24] Sixty-eight tumors highly expressed MYCN protein, and 81 were MYCN amplified. Thirty-nine tumors expressed MYC highly and were mutually exclusive of high MYCN expression. Segmental chromosomal aberrations were not examined in this study, except for MYCN amplification.[24] - Patients with favorable-histology (FH) tumors without high MYC/MYCN expression had favorable survival (3-year EFS, 89.7% ± 5.5%; 3-year OS, 97% ± 3.2%).
- Patients with undifferentiated or poorly differentiated histology tumors without MYC/MYCN expression had a 3-year EFS rate of 63.1% ± 13.6% and a 3-year OS rate of 83.5% ± 9.4%.
- Three-year EFS rates in patients with MYCN amplification, high MYCN expression, and high MYC expression were 48.1% ± 11.5%, 46.2% ± 12%, and 43.4% ± 23.1%, respectively, and OS rates were 65.8% ± 11.1%, 63.2% ± 12.1%, and 63.5% ± 19.2%, respectively.
- Further, when high expression of MYC and MYCN proteins were analyzed with other prognostic factors, including MYC/MYCN gene amplification, high MYC and MYCN protein expression was independent of other prognostic markers.
Most neuroblastomas with MYCN amplification in the International Neuroblastoma Pathology Classification system have unfavorable histology, but about 7% have FH. Of those with MYCN amplification and FH, most do not express MYCN, despite the gene being amplified, and have a more favorable prognosis than those that express MYCN.[25] Segmental chromosomal aberration at 11q is almost mutually exclusive of MYCN amplification. Neurotrophin receptor kinases Expression of neurotrophin receptor kinases and their ligands vary between high-risk and low-risk tumors. TrkA is found on low-risk tumors, and absence of its ligand NGF is postulated to lead to spontaneous tumor regression. In contrast, TrkB is found in high-risk tumors that also express its ligand, BDNF, which promotes neuroblastoma cell growth and survival.[26] Immune system inhibition Anti-GD2 antibodies, along with modulation of the immune system to enhance antineuroblastoma activity, are often used to help treat neuroblastoma. The anti-GD2 antibody (3F8), used for treating neuroblastoma exclusively at one institution, utilizes natural killer cells to kill the neuroblastoma cells. However, the natural killer cells can be inhibited by the interaction of HLA antigens and killer immunoglobulin receptor subtypes. Thus, the patient's immune system genes can help determine response to immunotherapy for neuroblastoma.[27,28] A report on the effects of immune system genes on response to dinutuximab, a commercially available anti-GD2 antibody, awaits publication. (Refer to the PDQ summary on Neuroblastoma Treatment for information about the treatment of neuroblastoma.) References:
-
Cohn SL, Pearson AD, London WB, et al.: The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 27 (2): 289-97, 2009.
-
Schleiermacher G, Mosseri V, London WB, et al.: Segmental chromosomal alterations have prognostic impact in neuroblastoma: a report from the INRG project. Br J Cancer 107 (8): 1418-22, 2012.
-
Janoueix-Lerosey I, Schleiermacher G, Michels E, et al.: Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol 27 (7): 1026-33, 2009.
-
Schleiermacher G, Michon J, Ribeiro A, et al.: Segmental chromosomal alterations lead to a higher risk of relapse in infants with MYCN-non-amplified localised unresectable/disseminated neuroblastoma (a SIOPEN collaborative study). Br J Cancer 105 (12): 1940-8, 2011.
-
Carén H, Kryh H, Nethander M, et al.: High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci U S A 107 (9): 4323-8, 2010.
-
Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, et al.: Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol 28 (19): 3122-30, 2010.
-
Defferrari R, Mazzocco K, Ambros IM, et al.: Influence of segmental chromosome abnormalities on survival in children over the age of 12 months with unresectable localised peripheral neuroblastic tumours without MYCN amplification. Br J Cancer 112 (2): 290-5, 2015.
-
Pugh TJ, Morozova O, Attiyeh EF, et al.: The genetic landscape of high-risk neuroblastoma. Nat Genet 45 (3): 279-84, 2013.
-
Ambros PF, Ambros IM, Brodeur GM, et al.: International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br J Cancer 100 (9): 1471-82, 2009.
-
Kreissman SG, Seeger RC, Matthay KK, et al.: Purged versus non-purged peripheral blood stem-cell transplantation for high-risk neuroblastoma (COG A3973): a randomised phase 3 trial. Lancet Oncol 14 (10): 999-1008, 2013.
-
Bagatell R, Beck-Popovic M, London WB, et al.: Significance of MYCN amplification in international neuroblastoma staging system stage 1 and 2 neuroblastoma: a report from the International Neuroblastoma Risk Group database. J Clin Oncol 27 (3): 365-70, 2009.
-
Peifer M, Hertwig F, Roels F, et al.: Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526 (7575): 700-4, 2015.
-
Valentijn LJ, Koster J, Zwijnenburg DA, et al.: TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet 47 (12): 1411-4, 2015.
-
Cheung NK, Zhang J, Lu C, et al.: Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 307 (10): 1062-71, 2012.
-
Molenaar JJ, Koster J, Zwijnenburg DA, et al.: Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483 (7391): 589-93, 2012.
-
Sausen M, Leary RJ, Jones S, et al.: Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet 45 (1): 12-7, 2013.
-
Bresler SC, Weiser DA, Huwe PJ, et al.: ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 26 (5): 682-94, 2014.
-
Janoueix-Lerosey I, Lequin D, Brugières L, et al.: Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455 (7215): 967-70, 2008.
-
Eleveld TF, Oldridge DA, Bernard V, et al.: Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet 47 (8): 864-71, 2015.
-
Schramm A, Köster J, Assenov Y, et al.: Mutational dynamics between primary and relapse neuroblastomas. Nat Genet 47 (8): 872-7, 2015.
-
Bellini A, Bernard V, Leroy Q, et al.: Deep Sequencing Reveals Occurrence of Subclonal ALK Mutations in Neuroblastoma at Diagnosis. Clin Cancer Res 21 (21): 4913-21, 2015.
-
Kurihara S, Hiyama E, Onitake Y, et al.: Clinical features of ATRX or DAXX mutated neuroblastoma. J Pediatr Surg 49 (12): 1835-8, 2014.
-
Mac SM, D'Cunha CA, Farnham PJ: Direct recruitment of N-myc to target gene promoters. Mol Carcinog 29 (2): 76-86, 2000.
-
Wang LL, Teshiba R, Ikegaki N, et al.: Augmented expression of MYC and/or MYCN protein defines highly aggressive MYC-driven neuroblastoma: a Children's Oncology Group study. Br J Cancer 113 (1): 57-63, 2015.
-
Suganuma R, Wang LL, Sano H, et al.: Peripheral neuroblastic tumors with genotype-phenotype discordance: a report from the Children's Oncology Group and the International Neuroblastoma Pathology Committee. Pediatr Blood Cancer 60 (3): 363-70, 2013.
-
Maris JM, Matthay KK: Molecular biology of neuroblastoma. J Clin Oncol 17 (7): 2264-79, 1999.
-
Forlenza CJ, Boudreau JE, Zheng J, et al.: KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma. J Clin Oncol 34 (21): 2443-51, 2016.
-
Venstrom JM, Zheng J, Noor N, et al.: KIR and HLA genotypes are associated with disease progression and survival following autologous hematopoietic stem cell transplantation for high-risk neuroblastoma. Clin Cancer Res 15 (23): 7330-4, 2009.
RetinoblastomaRetinoblastoma is a tumor that occurs in heritable (25%-30%) and nonheritable (70%-75%) forms. Heritable disease is defined by the presence of a germline mutation of the RB1 gene. This germline mutation may have been inherited from an affected progenitor (25% of cases) or may have occurred in a germ cell before conception or in utero during early embryogenesis in patients with sporadic disease (75% of cases). The presence of positive family history or bilateral or multifocal disease is suggestive of heritable disease. Heritable retinoblastoma may manifest as unilateral or bilateral disease. The penetrance of the RB1 mutation (laterality, age at diagnosis, and number of tumors) is probably dependent on concurrent genetic modifiers such as MDM2 and MDM4 polymorphisms.[1,2] All children with bilateral disease and approximately 15% of patients with unilateral disease are presumed to have the heritable form, even though only 25% have an affected parent. In heritable retinoblastoma, tumors tend to be diagnosed at a younger age than in the nonheritable form of the disease. Unilateral retinoblastoma in children younger than 1 year raises concern for heritable disease, whereas older children with a unilateral tumor are more likely to have the nonheritable form of the disease.[3] The genomic landscape of retinoblastoma is driven by alterations in RB1 that lead to biallelic inactivation.[4,5] A rare cause of RB1 inactivation is chromothripsis, which may be difficult to detect by conventional methods.[6] Other recurring genomic changes that occur in a small minority of tumors include BCOR mutation/deletion, MYCN amplification, and OTX2 amplification.[4,5,6] A study of 1,068 unilateral nonfamilial retinoblastoma tumors reported that a small percentage of cases (approximately 3%) lacked evidence of RB1 loss. Approximately one-half of these cases with no evidence of RB1 loss (representing approximately 1.5% of all unilateral nonfamilial retinoblastoma) showed MYCN amplification.[5] (Refer to the PDQ summary on Retinoblastoma Treatment for information about the treatment of retinoblastoma.) References:
-
Castéra L, Sabbagh A, Dehainault C, et al.: MDM2 as a modifier gene in retinoblastoma. J Natl Cancer Inst 102 (23): 1805-8, 2010.
-
de Oliveira Reis AH, de Carvalho IN, de Sousa Damasceno PB, et al.: Influence of MDM2 and MDM4 on development and survival in hereditary retinoblastoma. Pediatr Blood Cancer 59 (1): 39-43, 2012.
-
Zajaczek S, Jakubowska A, Kurzawski G, et al.: Age at diagnosis to discriminate those patients for whom constitutional DNA sequencing is appropriate in sporadic unilateral retinoblastoma. Eur J Cancer 34 (12): 1919-21, 1998.
-
Zhang J, Benavente CA, McEvoy J, et al.: A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481 (7381): 329-34, 2012.
-
Rushlow DE, Mol BM, Kennett JY, et al.: Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet Oncol 14 (4): 327-34, 2013.
-
McEvoy J, Nagahawatte P, Finkelstein D, et al.: RB1 gene inactivation by chromothripsis in human retinoblastoma. Oncotarget 5 (2): 438-50, 2014.
Kidney TumorsWilms Tumor Wilms tumor is thought to arise from clonal expansion of a nephrogenic rest. Mutations in many genes can perturb renal development and lead to cancer. This is in contrast to retinoblastoma, for example, in which a mutation in a single gene (RB1) is the oncogenic driver. Approximately one-third of Wilms tumor cases involve mutations in WT1, CTNNB1, or WTX.[1,2] Another subset of Wilms tumor cases result from mutations in miRNA processing genes (miRNAPG), including DROSHA, DGCR8, DICER1, and XPO5.[3,4,5,6] Other genes that are recurrently mutated in Wilms tumor include SIX1 and SIX2 (transcription factors that play key roles in early renal development) [3,4] and MLLT1 (a gene involved in transcriptional elongation in early development).[7] Anaplastic Wilms tumor is characterized by the presence of TP53 mutations. Elevated rates of Wilms tumor are observed in a number of genetic disorders, including WAGR (Wilms tumor, aniridia, genitourinary anomalies, and mental retardation) syndrome, Beckwith-Wiedemann syndrome, hemihypertrophy, Denys-Drash syndrome, and Perlman syndrome.[8] Other genetic causes that have been observed in familial Wilms tumor cases include germline mutations in REST and CTR9.[9,10] The genomic and genetic characteristics of Wilms tumor are summarized below. Wilms tumor 1gene (WT1) The WT1 gene is located on the short arm of chromosome 11 (11p13). WT1 is a transcription factor that is required for normal genitourinary development and is important for differentiation of the renal blastema.[11]WT1 mutations are observed in 10% to 20% of cases of sporadic Wilms tumor.[1,11,12] Wilms tumor with a WT1 mutation is characterized by the following: - Evidence of WNT pathway activation by activating mutations in the beta-catenin gene (CTNNB1) is common.[12,13,14]
- Loss of heterozygosity at 11p15 is commonly observed, as paternal uniparental disomy for chromosome 11 represents a common mechanism for losing the remaining normal WT1 allele.[12,15]
- Nephrogenic rests are benign foci of embryonal kidney cells that abnormally persist into postnatal life. Intralobar nephrogenic rests occur in approximately 20% of Wilms tumor cases. They are observed at high rates in cases with genetic syndromes that have WT1 mutations such as WAGR and Denys-Drash syndromes.[16] Intralobar nephrogenic rests are also observed in cases with sporadic WT1 and MLLT1 mutations.[7,17]
- WT1 germline mutations are uncommon (2%-4%) in nonsyndromic Wilms tumor.[18,19]
- WT1 mutations and 11p15 loss of heterozygosity were associated with relapse in patients with very low-risk Wilms tumor in one study of 56 patients who did not receive chemotherapy.[20] These findings need validation but may provide biomarkers for stratifying patients in the future.
Germline WT1 mutations are more common in children with Wilms tumor and one of the following: - WAGR syndrome, Denys-Drash syndrome,[21] or Frasier syndrome.[22]
- Genitourinary anomalies, including hypospadias and cryptorchidism.
- Bilateral Wilms tumor.
- Unilateral Wilms tumor with nephrogenic rests in the contralateral kidney.
- Stromal and rhabdomyomatous differentiation.
Syndromic conditions with germline WT1 mutations include WAGR syndrome, Denys-Drash syndrome,[21] and Frasier syndrome.[22] - WAGR syndrome. Children with WAGR syndrome (Wilms tumor, aniridia, genitourinary anomalies, and mental retardation) are at high risk (>30%) of developing Wilms tumor. WAGR syndrome results from deletions at chromosome 11p13 that involve a set of contiguous genes that includes the WT1 and PAX6 genes.
Inactivating mutations or deletions in the PAX6 gene lead to aniridia, while deletion of WT1 confers the increased risk of Wilms tumor. Sporadic aniridia in which WT1 is not deleted is not associated with increased risk of Wilms tumor. Accordingly, children with familial aniridia, generally occurring for many generations, and without renal abnormalities, have a normal WT1 gene and are not at an increased risk of Wilms tumor.[23,24] Wilms tumor in children with WAGR syndrome is characterized by an excess of bilateral disease, intralobar nephrogenic rests-associated favorable-histology (FH) tumors of mixed cell type, and early age at diagnosis.[25] The mental retardation in WAGR syndrome may be secondary to deletion of other genes, including SLC1A2 or BDNF.[26]
Germline WT1 point mutations produce genetic syndromes that are characterized by nephropathy, 46XY disorder of sex development, and varying risks of Wilms tumor.[27,28] - Denys-Drash and Frasier syndromes. Denys-Drash syndrome is characterized by nephrotic syndrome caused by diffuse mesangial sclerosis, XY pseudohermaphroditism, and increased risk of Wilms tumor. Frasier syndrome is characterized by progressive nephropathy caused by focal segmental glomerulosclerosis, gonadoblastoma, and XY pseudohermaphroditism.
WT1 mutations in Denys-Drash syndrome are most often missense mutations in exons 8 and 9, which code for the DNA binding region of WT1.[21] By contrast, WT1 mutations in Frasier syndrome typically occur in intron 9 at the KTS site, and they affect an alternative splicing, thereby preventing production of the usually more abundant WT1 +KTS isoform.[29] Studies evaluating genotype/phenotype correlations of WT1 mutations have shown that the risk of Wilms tumor is highest for truncating mutations (14 of 17 cases, 82%) and lower for missense mutations (27 of 67 cases, 42%). The risk is lowest for KTS splice site mutations (1 of 27 cases, 4%).[27,28] Bilateral Wilms tumor was more common in cases with WT1-truncating mutations (9 of 14 cases) than in cases with WT1 missense mutations (3 of 27 cases).[27,28] These genomic studies confirm previous estimates of elevated risk of Wilms tumor for children with Denys-Drash syndrome and low risk of Wilms tumor for children with Frasier syndrome.
Late effects associated with WAGR syndrome and Wilms tumor include the following: - Children with WAGR syndrome or other germline WT1 mutations are monitored throughout their lives because they are at increased risk of developing hypertension, nephropathy, and renal failure.[30]
- Patients with Wilms tumor and aniridia without genitourinary abnormalities are at lower risk but are monitored for nephropathy or renal failure.[31]
- Children with Wilms tumor and any genitourinary anomalies are also at increased risk of late renal failure and are monitored. Features associated with germline WT1 mutations that increase the risk of developing renal failure include the following:[30]
- Stromal predominant histology.
- Bilateral disease.
- Intralobar nephrogenic rests.
- Wilms tumor diagnosed before age 2 years.
(Refer to the Late effects after Wilms tumor therapy section of the PDQ summary on Wilms Tumor and Other Childhood Kidney Tumors Treatment for more information about the late effects associated with Wilms tumor.) Beta-cateningene (CTNNB1) Somatic activating mutations of the CTNNB1 gene have been reported to occur in 15% of patients with Wilms tumor.[2,12,14,32] These CTNNB1 mutations result in activation of the WNT pathway, which plays a prominent role in the developing kidney.[33]CTNNB1 mutations commonly occur with WT1 mutations, and most cases of Wilms tumor with WT1 mutations have a concurrent CTNNB1 mutation.[12,14,32] Activation of beta-catenin in the presence of intact WT1 protein appears to be inadequate to promote tumor development because CTNNB1 mutations are rarely found in the absence of a WT1 or WTX mutation, except when associated with a MLLT1 mutation.[2,34]CTNNB1 mutations appear to be late events in Wilms tumor development because they are found in tumors but not in nephrogenic rests.[17] Wilms tumor gene on the X chromosome (WTX) WTX, which is also called FAM123B, is located on the X chromosome at Xq11.1. It is altered in 15% to 20% of Wilms tumor cases.[35,1,2,12,36] Germline mutations in WTX cause an X-linked sclerosing bone dysplasia, osteopathia striata congenita with cranial sclerosis (MIM300373).[37] Despite having germline WTX mutations, individuals with osteopathia striata congenita are not predisposed to tumor development.[37] The WTX protein appears to be involved in both the degradation of beta-catenin and in the intracellular distribution of APC protein.[34,38]WTX is most commonly altered by deletions involving part or all of the WTX gene, with deleterious point mutations occurring less commonly.[1,12,35] Most Wilms tumor cases with WTX alterations have epigenetic 11p15 abnormalities.[12] WTX alterations are equally distributed between males and females, and WTX inactivation has no apparent effect on clinical presentation or prognosis.[1] Imprinting Cluster Regions (ICRs) on chromosome 11p15 (WT2) and Beckwith-Wiedemann syndrome A second Wilms tumor locus, WT2, maps to an imprinted region of chromosome 11p15.5; when it is a germline mutation, it causes Beckwith-Wiedemann syndrome. About 3% of children with Wilms tumor have germline epigenetic or genetic changes at the 11p15.5 growth regulatory locus without any clinical manifestations of overgrowth. Like children with Beckwith-Wiedemann syndrome, these children have an increased incidence of bilateral Wilms tumor or familial Wilms tumor.[26] Approximately 80% of patients with Beckwith-Wiedemann syndrome have a molecular defect of the 11p15 domain.[39] Various molecular mechanisms underlying Beckwith-Wiedemann syndrome have been identified. Some of these abnormalities are genetic (germline mutations of the maternal allele of CDKN1C, paternal uniparental isodisomy of 11p15, or duplication of part of the 11p15 domain) but are more frequently epigenetic (loss of methylation of the maternal ICR2/KvDMR1 or gain of methylation of the maternal ICR1).[26,40] Several candidate genes at the WT2 locus comprise the two independent imprinted domains IGF2/H19 and KIP2/LIT1.[40] Loss of heterozygosity, which exclusively affects the maternal chromosome, has the effect of upregulating paternally active genes and silencing maternally active ones. A loss or switch of the imprint for genes (change in methylation status) in this region has also been frequently observed and results in the same functional aberrations.[26,39,40] A relationship between epigenotype and phenotype has been shown in Beckwith-Wiedemann syndrome, with a different rate of cancer in Beckwith-Wiedemann syndrome according to the type of alteration of the 11p15 region.[41] The overall tumor risk in patients with Beckwith-Wiedemann syndrome has been estimated to be between 5% and 10%, with a risk between 1% (loss of imprinting at ICR2) and 30% (gain of methylation at ICR1 and paternal 11p15 isodisomy). Development of Wilms tumor has been reported in patients with only ICR1 gain of methylation, whereas other tumors such as neuroblastoma or hepatoblastoma were reported in patients with paternal 11p15 isodisomy.[42,43,44] Loss of imprinting or gene methylation is rarely found at other loci, supporting the specificity of loss of imprinting at 11p15.5.[45] Interestingly, Wilms tumor in Asian children is not associated with either nephrogenic rests or IGF2 loss of imprinting.[46] Approximately one-fifth of patients with Beckwith-Wiedemann syndrome who develop Wilms tumor present with bilateral disease, and metachronous bilateral disease is also observed.[23,47,48] The prevalence of Beckwith-Wiedemann syndrome is about 1% among children with Wilms tumor reported to the National Wilms Tumor Study (NWTS).[48,49] Other genes and chromosomal alterations Additional genes and chromosomal alterations that have been implicated in the pathogenesis and biology of Wilms tumorinclude the following: - 1q. Gain of chromosome 1q is associated with an inferior outcome and is the single most powerful predictor of outcome. In the presence of 1q gain, neither 1p nor 16q loss is significant.[50,51] Gain of chromosome 1q is one of the most common cytogenetic abnormalities in Wilms tumor and is observed in approximately 30% of tumors.
In an analysis of FH Wilms tumor from 1,114 patients from NWTS-5 (COG-Q9401/NCT00002611), 28% of the tumors displayed 1q gain.[50] - The 8-year event-free survival (EFS) rate was 77% for patients with 1q gain and 90% for those lacking 1q gain (P < .001). Within each disease stage, 1q gain was associated with inferior EFS.
- The 8-year overall survival (OS) rate was 88% for those with 1q gain and 96% for those lacking 1q gain (P < .001). OS was significantly inferior in cases with stage I disease (P < .0015) and stage IV disease (P = .011).
- 16q and 1p. Additional tumor-suppressor or tumor-progression genes may lie on chromosomes 16q and 1p, as evidenced by loss of heterozygosity for these regions in 17% and 11% of Wilms tumor cases, respectively.[52]
- In large NWTS studies, patients with tumor-specific loss of these loci had significantly worse relapse-free survival and OS rates. Combined loss of 1p and 16q are used to select FH Wilms tumor patients for more aggressive therapy in the current Children's Oncology Group (COG) study. However, a U.K. study of more than 400 patients found no significant association between 1p deletion and poor prognosis, but a poor prognosis was associated with 16q loss of heterozygosity.[53]
- An Italian study of 125 patients, using treatment quite similar to that in the COG study, found significantly worse prognosis in those with 1p deletions but not 16q deletions.[54]
These conflicting results may arise from the greater prognostic significance of 1q gain described above. Loss of heterozygosity of 16q and 1p loses significance as independent prognostic markers in the presence of 1q gain. However, in the absence of 1q gain, loss of heterozygosity of 16q and 1p retains their adverse prognostic impact.[50] The loss of heterozygosity of 16q and 1p appears to arise from complex chromosomal events that result in 1q loss of heterozygosity or 1q gain. The change in 1q appears to be the critical tumorigenic genetic event.[55] - miRNAPG. Mutations in selected miRNAPG are observed in approximately 20% of Wilms tumor cases.[3,4,5,6] The products of these genes direct the maturation of miRNAs from the initial pri-miRNA transcripts to functional cytoplasmic miRNAs (refer to Figure 13).[56] The most commonly mutated miRNAPG is DROSHA, with a recurrent mutation (E1147K) affecting a metal-binding residue of the RNase IIIb domain, representing about 80% of DROSHA-mutated tumors. Other miRNAPG that are mutated in Wilms tumor include DGCR8, DICER1, TARBP2, DIS3L2, and XPO5. These mutations are generally mutually exclusive, and they appear to be deleterious and result in impaired expression of tumor-suppressing miRNAs. A striking sex bias was noted in mutations for DGCR8 (located on chromosome 22q11), with 38 of 43 cases (88%) arising in girls.[3,4]
Germline mutations in miRNAPG are observed for DICER1 and DIS3L2, with mutations in the former causing DICER1 syndrome and mutations in the latter causing Perlman syndrome. - DICER1 syndrome is typically caused by inherited truncating mutations in DICER1, with tumor formation following acquisition of a missense mutation in a domain of the remaining allele of DICER1 (the RNase IIIb domain) responsible for processing miRNAs derived from the 5p arms of pre-miRNAs.[57] Tumors associated with DICER1 syndrome include pleuropulmonary blastoma, cystic nephroma, ovarian sex cord-stromal tumors, multinodular goiter, and embryonal rhabdomyosarcoma.[57] Wilms tumor is an uncommon presentation of the DICER1 syndrome. In one study, three families with DICER1 syndrome included children with Wilms tumor, with two of the Wilms tumor cases showing the typical second DICER1 mutation in the RNase IIIb domain.[58] Another study identified DICER1 mutations in 2 of 48 familial Wilms tumor families.[59] Large sequencing studies of Wilms tumor cohorts have also observed occasional cases with DICER1 mutations.[4,5]
- Perlman syndrome is a rare overgrowth disorder caused by mutations in DIS3L2, which encodes a ribonuclease that is responsible for degrading pre-let-7 miRNA.[60,61] The prognosis of Perlman syndrome is poor, with a high neonatal mortality rate. In a survey of published cases of Perlman syndrome (N = 28), in infants who survived beyond the neonatal period, approximately two-thirds developed Wilms tumor, and all patients showed developmental delay. Fetal macrosomia, ascites, and polyhydramnios are frequent manifestations.[62]
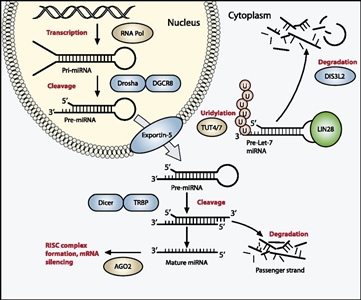
Figure 13. The miRNA processing pathway is commonly mutated in Wilms tumor. Expression of mature miRNA is initiated by RNA polymerase-mediated transcription of DNA-encoded sequences into pri-miRNA, which form a long double-stranded hairpin. This structure is then cleaved by a complex of Drosha and DGCR8 into a smaller pre-miRNA hairpin, which is exported from the nucleus and then cleaved by Dicer (an RNase) and TRBP (with specificity for dsRNA) to remove the hairpin loop and leave two single-stranded miRNAs. The functional strand binds to Argonaute (Ago2) proteins into the RNA-induced silencing complex (RISC), where it guides the complex to its target mRNA, while the nonfunctional strand is degraded. Targeting of mRNAs by this method results in mRNA silencing by mRNA cleavage, translational repression, or deadenylation. Let-7 miRNAs are a family of miRNAs highly expressed in ESCs with tumor suppressor properties. In cases in which LIN28 is overexpressed, LIN28 binds to pre-Let-7 miRNA, preventing DICER from binding and resulting in LIN28-activated polyuridylation by TUT4 or TUT7, causing reciprocal DIS3L2-mediated degradation of Let-7 pre-miRNAs. Genes involved in miRNA processing that have been associated with Wilms' tumor are highlighted in blue (inactivating) and green (activating) and include DROSHA, DGCR8, XPO5 (encoding exportin-5), DICER1, TARBP2, DIS3L2, and LIN28. Copyright © 2015 Hohenstein et al.; Published by Cold Spring Harbor Laboratory Press. Genes Dev. 2015 Mar 1; 29(5): 467-482. doi: 10.1101/gad.256396.114. This article is distributed exclusively by Cold Spring Harbor Laboratory Press under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.
- SIX1 and SIX2. SIX1 and SIX2 are highly homologous transcription factors that play key roles in early renal development and are expressed in the metanephric mesenchyme, where they maintain the mesenchymal progenitor population. The frequency of SIX1 mutations is 3% to 4% in Wilms tumor, and the frequency of SIX2 mutations in Wilms tumor is 1% to 3%.[3,4] Virtually all SIX1 and SIX2 mutations are in exon 1 and result in a glutamine-to-arginine mutation at position 177. Mutations in WT1, WTX, and CTNNB1 are infrequent in cases with SIX1/SIX2 or miRNAPG mutations. Conversely, SIX1/SIX2 mutations and miRNAPG mutations tend to occur together.
- MLLT1. Approximately 4% of Wilms tumor cases have mutations in the highly conserved YEATS domain of MLLT1 (ENL), a gene known to be involved in transcriptional elongation by RNA polymerase II during early development.[7] The mutant MLLT1 protein shows altered binding to acetylated histone tails. Patients with MLLT1-mutant tumors present at a younger age and have a high prevalence of precursor intralobar nephrogenic rests, supporting a model whereby activating MLLT1 mutations early in renal development result in the development of Wilms tumor.
- TP53 (tumor suppressor gene). Most anaplastic Wilms tumor cases show mutations in the TP53 tumor suppressor gene.[63,64,65]TP53 may be useful as an unfavorable prognostic marker.[63,64] A study of 40 patients with diffuse anaplastic Wilms tumor identified 25 patients with TP53 alterations (22 with TP53 mutations with or without 17p loss, and 3 with only 17p loss). The 25 cases with TP53 alterations had a significantly lower EFS and OS than did those without TP53 alterations.[65] Microdissection of focally anaplastic Wilms tumor demonstrated TP53 mutations in anaplastic but not nonanaplastic areas of the tumor, suggesting that acquisition of TP53 mutation may be inherent in the process of becoming anaplastic.[66]
- FBXW7. FBXW7, a ubiquitin ligase component, is a gene that has been identified as recurrently mutated at low rates in Wilms tumor. Mutations of this gene have been associated with epithelial-type tumor histology.[67]
- 9q22.3 microdeletion syndrome. Patients with 9q22.3 microdeletion syndrome have an increased risk of Wilms tumor.[68,69] The chromosomal region with germline deletion includes PTCH1, the gene that is mutated in Gorlin syndrome (nevoid basal cell carcinoma syndrome associated with osteosarcoma). 9q22.3 microdeletion syndrome is characterized by the clinical findings of Gorlin syndrome, as well as developmental delay and/or intellectual disability, metopic craniosynostosis, obstructive hydrocephalus, prenatal and postnatal macrosomia, and seizures.[68] Five patients who presented with Wilms tumor in the context of a constitutional 9q22.3 microdeletion have been reported.[69,70,71]
- MYCN. MYCN copy number gain was observed in approximately 13% of Wilms tumor cases, and it was more common in anaplastic cases (7 of 23 cases, 30%) than in nonanaplastic cases (11.2%).[72] Activating mutations at codon 44 (p.P44L) were identified in approximately 4% of Wilms tumor cases.[72] Germline copy number gain at MYCN has been reported in a bilateral Wilms tumor case, and germline MYCN duplication was also reported for a child with prenatal bilateral nephroblastomatosis and a family history of nephroblastoma.[73]
- CTR9. Inactivating CTR9 germline mutations were identified in 3 of 35 familial Wilms tumor pedigrees.[10]CTR9, which is located at chromosome 11p15.3, is a key component of the polymerase-associated factor 1 complex (PAF1c), which has multiple roles in RNA polymerase II regulation and is implicated in embryonic organogenesis and maintenance of embryonic stem cell pluripotency.
- REST. Inactivating germline mutations in REST (encoding RE1-silencing transcription factor) were identified in four familial Wilms tumor pedigrees.[9] REST is a transcriptional repressor that functions in cellular differentiation and embryonic development. Most REST mutations clustered within the portion of REST encoding the DNA-binding domain, and functional analyses showed that these mutations compromise REST transcriptional repression. When screened for REST mutations, 9 of 519 individuals with Wilms tumor who had no history of relatives with the disease tested positive for the mutation; some had parents who also tested positive.[9] These observations indicate that REST is a Wilms tumor predisposition gene associated with approximately 2% of Wilms tumor.
Figure 14 summarizes the genomic landscape of a selected cohort of Wilms tumor patients selected because they experienced relapse despite showing FH.[7] The 75 FH Wilms tumor cases were clustered by unsupervised analysis of gene expression data, resulting in six clusters. Five of six MLLT1-mutant tumors with available gene expression data were in cluster 3, and two were accompanied by CTNNB1 mutations. This cluster also contained four tumors with a mutation or small segment deletion of WT1, all of which also had either a mutation of CTNNB1 or small segment deletion or mutation of WTX. It also contained a substantial number of tumors with retention of imprinting of 11p15 (including all MLLT1-mutant tumors). The miRNAPG-mutated cases clustered together and were mutually exclusive with both MLLT1 and with WT1/WTX/CTNNB1-mutated cases.
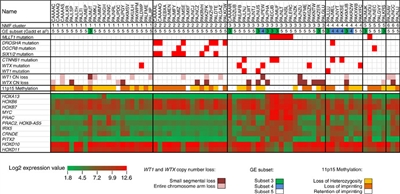
Figure 14. Unsupervised analysis of gene expression data. Non-negative Matrix Factorization (NMF) analysis of 75 FH Wilms tumor resulted in six clusters. Five of six MLLT1 mutant tumors with available gene expression data occurred in NMF cluster 3, and two were accompanied by CTNNB1 mutations. This cluster also contained a substantial number of tumors with retention of imprinting of 11p15 (including all MLLT1-mutant tumors), in contrast to other clusters, where most cases showed 11p15 loss of heterozygosity or retention of imprinting. Almost all miRNAPG-mutated cases were in NMF cluster 2, and most WT1, WTX, and CTNNB1 mutations were in NMF clusters 3 and 4. Copyright © 2015 Perlman, E. J. et al. MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology wilms tumours. Nat. Commun. 6:10013 doi: 10.1038/ncomms10013 (2015). This article is distributed by Nature Publishing Group, a division of Macmillan Publishers Limited under a Creative Commons Attribution 4.0 International License, as described at http://creativecommons.org/licenses/by/4.0/.
Renal Cell Carcinoma Translocation-positive carcinomas of the kidney are recognized as a distinct form of renal cell carcinoma (RCC) and may be the most common form of RCC in children, accounting for 40% to 50% of pediatric RCC.[74] In a Children's Oncology Group (COG) prospective clinical trial of 120 childhood and adolescent patients with RCC, nearly one-half of patients had translocation-positive RCC.[75] These carcinomas are characterized by translocations involving the transcription factor E3 gene (TFE3) located on Xp11.2. The TFE3 gene may partner with one of the following genes: - ASPSCR in t(X;17)(p11.2;q25).
- PRCC in t(X;1)(p11.2;q21).
- SFPQ in t(X;1)(p11.2;p34).
- NONO in inv(X;p11.2;q12).
- Clathrin heavy chain (CLTC) in t(X;17)(p11;q23).
Another less-common translocation subtype, t(6;11)(p21;q12), involving an Alpha-transcription factor EB (TFEB) gene fusion, induces overexpression of TFEB. The translocations involving TFE3 and TFEB induce overexpression of these proteins, which can be identified by immunohistochemistry.[76] Previous exposure to chemotherapy is the only known risk factor for the development of Xp11 translocation RCCs. In one study, the postchemotherapy interval ranged from 4 to 13 years. All reported patients received either a DNA topoisomerase II inhibitor and/or an alkylating agent.[77,78] Controversy exists as to the biological behavior of translocation RCC in children and young adults. Whereas some series have suggested a good prognosis when RCC is treated with surgery alone despite presenting at a more advanced stage (III/IV) than TFE-RCC, a meta-analysis reported that these patients have poorer outcomes.[79,80,81] The outcomes for these patients are being studied in the ongoing COG AREN03B2 (NCT00898365) biology and classification study. Vascular endothelial growth factor receptor-targeted therapies and mammalian target of rapamycin (mTOR) inhibitors seem to be active in Xp11 translocation metastatic RCC.[82] Recurrences have been reported 20 to 30 years after initial resection of the translocation-associated RCC.[83] Diagnosis of Xp11 translocation RCC needs to be confirmed by a molecular genetic approach, rather than using TFE3 immunohistochemistry alone, because reported cases have lacked the translocation. There is a rare subset of RCC cases that is positive for TFE3 and lack a TFE3 translocation, showing an ALK translocation instead. This subset of cases represents a newly recognized subgroup within RCC that is estimated to involve 15% to 20% of unclassified pediatric RCC. In the eight reported cases in children aged 6 to 16 years, the following was observed:[84,85,86,87] - ALK was fused to VCL (vinculin) in a t(2;10)(p23;q22) translocation (n = 3). The VCL translocation cases all occurred in children with sickle cell trait, whereas none of the TMP3 translocation cases did.
- ALK was fused to TPM3 (tropomyosin 3) (n = 3).
- ALK was fused to HOOK-1 on 1p32 (n = 1).
- t(1;2) translocation fusing ALK and TMP3 (n = 1).
(Refer to the PDQ summary on Wilms Tumor and Other Childhood Kidney Tumors Treatment for information about the treatment of renal cell carcinoma.) Rhabdoid Tumors of the Kidney Rhabdoid tumors in all anatomical locations have a common genetic abnormality-loss of function of the SMARCB1/INI1/SNF5/BAF47 gene located at chromosome 22q11.2. The following text refers to rhabdoid tumors without regard to their primary site. SMARCB1 encodes a component of the SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeling complex that has an important role in controlling gene transcription.[88,89] Loss of function occurs by deletions that lead to loss of part or all of the SMARCB1 gene and by mutations that are commonly frameshift or nonsense mutations that lead to premature truncation of the SMARCB1 protein.[89,90] A small percentage of rhabdoid tumors are caused by alterations in SMARCA4, which is the primary ATPase in the SWI/SNF complex.[91,92] Exome sequencing of 35 cases of rhabdoid tumor identified a very low mutation rate, with no genes having recurring mutations other than SMARCB1, which appeared to contribute to tumorigenesis.[93] Germline mutations of SMARCB1 have been documented in patients with one or more primary tumors of the brain and/or kidney, consistent with a genetic predisposition to the development of rhabdoid tumors.[94,95] Approximately one-third of patients with rhabdoid tumors have germline SMARCB1 alterations.[89,96] In most cases, the mutations are de novo and not inherited. The median age at diagnosis of children with rhabdoid tumors and a germline mutation or deletion is younger (6 months) than that of children with apparently sporadic disease (18 months).[97] Germline mosaicism has been suggested for several families with multiple affected siblings. It appears that patients with germline mutations may have the worst prognosis.[98,99] Germline mutations in SMARCA4 have also been reported in patients with rhabdoid tumors.[91,100] (Refer to the PDQ summary on Wilms Tumor and Other Childhood Kidney Tumors Treatment for information about the treatment of rhabdoid tumor of the kidney.) Clear Cell Sarcoma of the Kidney Clear cell sarcoma of the kidney is an uncommon renal tumor that comprises approximately 5% of all primary renal malignancies in children, and is observed most often before age 3 years. The molecular background of clear cell sarcoma of the kidney is poorly understood due to its rarity and lack of experimental models. Several biological features of clear cell sarcoma of the kidney have been described, including the following: - Internal tandem duplications in exon 15 of the BCOR gene (BCL6 corepressor) were reported in 100% (20 of 20 cases) of clear cell sarcoma of the kidney cases but in none of the other pediatric renal tumors evaluated.[101] Other reports have confirmed the finding of BCOR internal tandem duplications in clear cell sarcoma of the kidney.[102,103,104] Hence, BCOR internal tandem duplications appear to play a key role in the tumorigenesis of clear cell sarcoma of the kidney, and their identification should aid in the differential diagnosis of renal tumors.[101]
- The YWHAE-NUTM2 fusion (involving either NUTM2B or NUTM2E) resulting from t(10;17) was reported in 12% of cases of clear cell sarcoma of the kidney.[105] The presence of the YWHAE-NUTM2 fusion appears to be mutually exclusive with the presence of BCOR internal tandem duplications; this observation is based on a study of 22 cases of clear cell sarcoma of the kidney that included two cases with the YWHAE-NUTM2 fusion and 20 cases with BCOR internal tandem duplications.[102] The gene expression profiles for cases with the YWHAE-NUTM2 fusion were distinctive from those with BCOR internal tandem duplications.
- Evaluation of 13 clear cell sarcoma of the kidney tumors for changes in chromosome copy number, mutations, and rearrangements found a single case with the YWHAE-NUTM2 fusion and 12 cases with BCOR internal tandem duplications.[104,106] No other recurrent segmental chromosomal copy number changes or somatic variants (single nucleotide or small insertion/deletion) were identified, providing further support for the role of BCOR internal tandem duplication as the primary oncogenic driver for clear cell sarcoma of the kidney.[106]
(Refer to the PDQ summary on Wilms Tumor and Other Childhood Kidney Tumors Treatment for information about the treatment of clear cell tumor of the kidney.) References:
-
Wegert J, Wittmann S, Leuschner I, et al.: WTX inactivation is a frequent, but late event in Wilms tumors without apparent clinical impact. Genes Chromosomes Cancer 48 (12): 1102-11, 2009.
-
Ruteshouser EC, Robinson SM, Huff V: Wilms tumor genetics: mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer 47 (6): 461-70, 2008.
-
Walz AL, Ooms A, Gadd S, et al.: Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 27 (2): 286-97, 2015.
-
Wegert J, Ishaque N, Vardapour R, et al.: Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 27 (2): 298-311, 2015.
-
Rakheja D, Chen KS, Liu Y, et al.: Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun 2: 4802, 2014.
-
Torrezan GT, Ferreira EN, Nakahata AM, et al.: Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun 5: 4039, 2014.
-
Perlman EJ, Gadd S, Arold ST, et al.: MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology Wilms tumours. Nat Commun 6: 10013, 2015.
-
Dome JS, Huff V: Wilms Tumor Predisposition. In: Pagon RA, Adam MP, Bird TD, et al., eds.: GeneReviews. Seattle, WA: University of Washington, 2013, pp. Available online. Last accessed February 03, 2017.
-
Mahamdallie SS, Hanks S, Karlin KL, et al.: Mutations in the transcriptional repressor REST predispose to Wilms tumor. Nat Genet 47 (12): 1471-4, 2015.
-
Hanks S, Perdeaux ER, Seal S, et al.: Germline mutations in the PAF1 complex gene CTR9 predispose to Wilms tumour. Nat Commun 5: 4398, 2014.
-
Huff V: Wilms tumor genetics. Am J Med Genet 79 (4): 260-7, 1998.
-
Scott RH, Murray A, Baskcomb L, et al.: Stratification of Wilms tumor by genetic and epigenetic analysis. Oncotarget 3 (3): 327-35, 2012.
-
Corbin M, de Reyniès A, Rickman DS, et al.: WNT/beta-catenin pathway activation in Wilms tumors: a unifying mechanism with multiple entries? Genes Chromosomes Cancer 48 (9): 816-27, 2009.
-
Maiti S, Alam R, Amos CI, et al.: Frequent association of beta-catenin and WT1 mutations in Wilms tumors. Cancer Res 60 (22): 6288-92, 2000.
-
Gadd S, Huff V, Huang CC, et al.: Clinically relevant subsets identified by gene expression patterns support a revised ontogenic model of Wilms tumor: a Children's Oncology Group Study. Neoplasia 14 (8): 742-56, 2012.
-
Breslow NE, Beckwith JB, Perlman EJ, et al.: Age distributions, birth weights, nephrogenic rests, and heterogeneity in the pathogenesis of Wilms tumor. Pediatr Blood Cancer 47 (3): 260-7, 2006.
-
Fukuzawa R, Heathcott RW, More HE, et al.: Sequential WT1 and CTNNB1 mutations and alterations of beta-catenin localisation in intralobar nephrogenic rests and associated Wilms tumours: two case studies. J Clin Pathol 60 (9): 1013-6, 2007.
-
Diller L, Ghahremani M, Morgan J, et al.: Constitutional WT1 mutations in Wilms' tumor patients. J Clin Oncol 16 (11): 3634-40, 1998.
-
Little SE, Hanks SP, King-Underwood L, et al.: Frequency and heritability of WT1 mutations in nonsyndromic Wilms' tumor patients: a UK Children's Cancer Study Group Study. J Clin Oncol 22 (20): 4140-6, 2004.
-
Perlman EJ, Grundy PE, Anderson JR, et al.: WT1 mutation and 11P15 loss of heterozygosity predict relapse in very low-risk wilms tumors treated with surgery alone: a children's oncology group study. J Clin Oncol 29 (6): 698-703, 2011.
-
Pelletier J, Bruening W, Kashtan CE, et al.: Germline mutations in the Wilms' tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell 67 (2): 437-47, 1991.
-
Barbosa AS, Hadjiathanasiou CG, Theodoridis C, et al.: The same mutation affecting the splicing of WT1 gene is present on Frasier syndrome patients with or without Wilms' tumor. Hum Mutat 13 (2): 146-53, 1999.
-
Green DM, Breslow NE, Beckwith JB, et al.: Screening of children with hemihypertrophy, aniridia, and Beckwith-Wiedemann syndrome in patients with Wilms tumor: a report from the National Wilms Tumor Study. Med Pediatr Oncol 21 (3): 188-92, 1993.
-
Scott RH, Walker L, Olsen ØE, et al.: Surveillance for Wilms tumour in at-risk children: pragmatic recommendations for best practice. Arch Dis Child 91 (12): 995-9, 2006.
-
Breslow NE, Norris R, Norkool PA, et al.: Characteristics and outcomes of children with the Wilms tumor-Aniridia syndrome: a report from the National Wilms Tumor Study Group. J Clin Oncol 21 (24): 4579-85, 2003.
-
Scott RH, Douglas J, Baskcomb L, et al.: Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nat Genet 40 (11): 1329-34, 2008.
-
Lipska BS, Ranchin B, Iatropoulos P, et al.: Genotype-phenotype associations in WT1 glomerulopathy. Kidney Int 85 (5): 1169-78, 2014.
-
Lehnhardt A, Karnatz C, Ahlenstiel-Grunow T, et al.: Clinical and molecular characterization of patients with heterozygous mutations in wilms tumor suppressor gene 1. Clin J Am Soc Nephrol 10 (5): 825-31, 2015.
-
Barbaux S, Niaudet P, Gubler MC, et al.: Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat Genet 17 (4): 467-70, 1997.
-
Lange J, Peterson SM, Takashima JR, et al.: Risk factors for end stage renal disease in non-WT1-syndromic Wilms tumor. J Urol 186 (2): 378-86, 2011.
-
Breslow NE, Takashima JR, Ritchey ML, et al.: Renal failure in the Denys-Drash and Wilms' tumor-aniridia syndromes. Cancer Res 60 (15): 4030-2, 2000.
-
Koesters R, Ridder R, Kopp-Schneider A, et al.: Mutational activation of the beta-catenin proto-oncogene is a common event in the development of Wilms' tumors. Cancer Res 59 (16): 3880-2, 1999.
-
Koesters R, Niggli F, von Knebel Doeberitz M, et al.: Nuclear accumulation of beta-catenin protein in Wilms' tumours. J Pathol 199 (1): 68-76, 2003.
-
Major MB, Camp ND, Berndt JD, et al.: Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science 316 (5827): 1043-6, 2007.
-
Rivera MN, Kim WJ, Wells J, et al.: An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science 315 (5812): 642-5, 2007.
-
Fukuzawa R, Anaka MR, Weeks RJ, et al.: Canonical WNT signalling determines lineage specificity in Wilms tumour. Oncogene 28 (8): 1063-75, 2009.
-
Jenkins ZA, van Kogelenberg M, Morgan T, et al.: Germline mutations in WTX cause a sclerosing skeletal dysplasia but do not predispose to tumorigenesis. Nat Genet 41 (1): 95-100, 2009.
-
Grohmann A, Tanneberger K, Alzner A, et al.: AMER1 regulates the distribution of the tumor suppressor APC between microtubules and the plasma membrane. J Cell Sci 120 (Pt 21): 3738-47, 2007.
-
Satoh Y, Nakadate H, Nakagawachi T, et al.: Genetic and epigenetic alterations on the short arm of chromosome 11 are involved in a majority of sporadic Wilms' tumours. Br J Cancer 95 (4): 541-7, 2006.
-
Algar EM, St Heaps L, Darmanian A, et al.: Paternally inherited submicroscopic duplication at 11p15.5 implicates insulin-like growth factor II in overgrowth and Wilms' tumorigenesis. Cancer Res 67 (5): 2360-5, 2007.
-
Lennerz JK, Timmerman RJ, Grange DK, et al.: Addition of H19 'loss of methylation testing' for Beckwith-Wiedemann syndrome (BWS) increases the diagnostic yield. J Mol Diagn 12 (5): 576-88, 2010.
-
Bliek J, Gicquel C, Maas S, et al.: Epigenotyping as a tool for the prediction of tumor risk and tumor type in patients with Beckwith-Wiedemann syndrome (BWS). J Pediatr 145 (6): 796-9, 2004.
-
Rump P, Zeegers MP, van Essen AJ: Tumor risk in Beckwith-Wiedemann syndrome: A review and meta-analysis. Am J Med Genet A 136 (1): 95-104, 2005.
-
Brioude F, Lacoste A, Netchine I, et al.: Beckwith-Wiedemann syndrome: growth pattern and tumor risk according to molecular mechanism, and guidelines for tumor surveillance. Horm Res Paediatr 80 (6): 457-65, 2013.
-
Bjornsson HT, Brown LJ, Fallin MD, et al.: Epigenetic specificity of loss of imprinting of the IGF2 gene in Wilms tumors. J Natl Cancer Inst 99 (16): 1270-3, 2007.
-
Fukuzawa R, Breslow NE, Morison IM, et al.: Epigenetic differences between Wilms' tumours in white and east-Asian children. Lancet 363 (9407): 446-51, 2004.
-
DeBaun MR, Siegel MJ, Choyke PL: Nephromegaly in infancy and early childhood: a risk factor for Wilms tumor in Beckwith-Wiedemann syndrome. J Pediatr 132 (3 Pt 1): 401-4, 1998.
-
DeBaun MR, Tucker MA: Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J Pediatr 132 (3 Pt 1): 398-400, 1998.
-
Breslow N, Olshan A, Beckwith JB, et al.: Epidemiology of Wilms tumor. Med Pediatr Oncol 21 (3): 172-81, 1993.
-
Gratias EJ, Dome JS, Jennings LJ, et al.: Association of Chromosome 1q Gain With Inferior Survival in Favorable-Histology Wilms Tumor: A Report From the Children's Oncology Group. J Clin Oncol 34 (26): 3189-94, 2016.
-
Chagtai T, Zill C, Dainese L, et al.: Gain of 1q As a Prognostic Biomarker in Wilms Tumors (WTs) Treated With Preoperative Chemotherapy in the International Society of Paediatric Oncology (SIOP) WT 2001 Trial: A SIOP Renal Tumours Biology Consortium Study. J Clin Oncol 34 (26): 3195-203, 2016.
-
Grundy PE, Breslow NE, Li S, et al.: Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: a report from the National Wilms Tumor Study Group. J Clin Oncol 23 (29): 7312-21, 2005.
-
Messahel B, Williams R, Ridolfi A, et al.: Allele loss at 16q defines poorer prognosis Wilms tumour irrespective of treatment approach in the UKW1-3 clinical trials: a Children's Cancer and Leukaemia Group (CCLG) Study. Eur J Cancer 45 (5): 819-26, 2009.
-
Spreafico F, Gamba B, Mariani L, et al.: Loss of heterozygosity analysis at different chromosome regions in Wilms tumor confirms 1p allelic loss as a marker of worse prognosis: a study from the Italian Association of Pediatric Hematology and Oncology. J Urol 189 (1): 260-6, 2013.
-
Gratias EJ, Jennings LJ, Anderson JR, et al.: Gain of 1q is associated with inferior event-free and overall survival in patients with favorable histology Wilms tumor: a report from the Children's Oncology Group. Cancer 119 (21): 3887-94, 2013.
-
Hohenstein P, Pritchard-Jones K, Charlton J: The yin and yang of kidney development and Wilms' tumors. Genes Dev 29 (5): 467-82, 2015.
-
Foulkes WD, Priest JR, Duchaine TF: DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer 14 (10): 662-72, 2014.
-
Wu MK, Sabbaghian N, Xu B, et al.: Biallelic DICER1 mutations occur in Wilms tumours. J Pathol 230 (2): 154-64, 2013.
-
Palculict TB, Ruteshouser EC, Fan Y, et al.: Identification of germline DICER1 mutations and loss of heterozygosity in familial Wilms tumour. J Med Genet 53 (6): 385-8, 2016.
-
Astuti D, Morris MR, Cooper WN, et al.: Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44 (3): 277-84, 2012.
-
Chang HM, Triboulet R, Thornton JE, et al.: A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497 (7448): 244-8, 2013.
-
Alessandri JL, Cuillier F, Ramful D, et al.: Perlman syndrome: report, prenatal findings and review. Am J Med Genet A 146A (19): 2532-7, 2008.
-
Bardeesy N, Falkoff D, Petruzzi MJ, et al.: Anaplastic Wilms' tumour, a subtype displaying poor prognosis, harbours p53 gene mutations. Nat Genet 7 (1): 91-7, 1994.
-
el Bahtimi R, Hazen-Martin DJ, Re GG, et al.: Immunophenotype, mRNA expression, and gene structure of p53 in Wilms' tumors. Mod Pathol 9 (3): 238-44, 1996.
-
Wallkamm V, Dörlich R, Rahm K, et al.: Live imaging of Xwnt5A-ROR2 complexes. PLoS One 9 (10): e109428, 2014.
-
Bardeesy N, Beckwith JB, Pelletier J: Clonal expansion and attenuated apoptosis in Wilms' tumors are associated with p53 gene mutations. Cancer Res 55 (2): 215-9, 1995.
-
Williams RD, Al-Saadi R, Chagtai T, et al.: Subtype-specific FBXW7 mutation and MYCN copy number gain in Wilms' tumor. Clin Cancer Res 16 (7): 2036-45, 2010.
-
Muller E, Hudgins L: 9q22.3 Microdeletion. In: Pagon RA, Adam MP, Bird TD, et al., eds.: GeneReviews. Seattle, WA: University of Washington, 2013, pp. Available online. Last accessed February 03, 2017.
-
Isidor B, Bourdeaut F, Lafon D, et al.: Wilms' tumor in patients with 9q22.3 microdeletion syndrome suggests a role for PTCH1 in nephroblastomas. Eur J Hum Genet 21 (7): 784-7, 2013.
-
Garavelli L, Piemontese MR, Cavazza A, et al.: Multiple tumor types including leiomyoma and Wilms tumor in a patient with Gorlin syndrome due to 9q22.3 microdeletion encompassing the PTCH1 and FANC-C loci. Am J Med Genet A 161A (11): 2894-901, 2013.
-
Cajaiba MM, Bale AE, Alvarez-Franco M, et al.: Rhabdomyosarcoma, Wilms tumor, and deletion of the patched gene in Gorlin syndrome. Nat Clin Pract Oncol 3 (10): 575-80, 2006.
-
Williams RD, Chagtai T, Alcaide-German M, et al.: Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget 6 (9): 7232-43, 2015.
-
Fievet A, Belaud-Rotureau MA, Dugay F, et al.: Involvement of germline DDX1-MYCN duplication in inherited nephroblastoma. Eur J Med Genet 56 (12): 643-7, 2013.
-
Geller JI, Dome JS: Local lymph node involvement does not predict poor outcome in pediatric renal cell carcinoma. Cancer 101 (7): 1575-83, 2004.
-
Geller JI, Ehrlich PF, Cost NG, et al.: Characterization of adolescent and pediatric renal cell carcinoma: A report from the Children's Oncology Group study AREN03B2. Cancer 121 (14): 2457-64, 2015.
-
Argani P, Hicks J, De Marzo AM, et al.: Xp11 translocation renal cell carcinoma (RCC): extended immunohistochemical profile emphasizing novel RCC markers. Am J Surg Pathol 34 (9): 1295-303, 2010.
-
Argani P, Laé M, Ballard ET, et al.: Translocation carcinomas of the kidney after chemotherapy in childhood. J Clin Oncol 24 (10): 1529-34, 2006.
-
Ramphal R, Pappo A, Zielenska M, et al.: Pediatric renal cell carcinoma: clinical, pathologic, and molecular abnormalities associated with the members of the mit transcription factor family. Am J Clin Pathol 126 (3): 349-64, 2006.
-
Geller JI, Argani P, Adeniran A, et al.: Translocation renal cell carcinoma: lack of negative impact due to lymph node spread. Cancer 112 (7): 1607-16, 2008.
-
Camparo P, Vasiliu V, Molinie V, et al.: Renal translocation carcinomas: clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol 32 (5): 656-70, 2008.
-
Qiu Rao, Bing Guan, Zhou XJ: Xp11.2 Translocation renal cell carcinomas have a poorer prognosis than non-Xp11.2 translocation carcinomas in children and young adults: a meta-analysis. Int J Surg Pathol 18 (6): 458-64, 2010.
-
Malouf GG, Camparo P, Oudard S, et al.: Targeted agents in metastatic Xp11 translocation/TFE3 gene fusion renal cell carcinoma (RCC): a report from the Juvenile RCC Network. Ann Oncol 21 (9): 1834-8, 2010.
-
Rais-Bahrami S, Drabick JJ, De Marzo AM, et al.: Xp11 translocation renal cell carcinoma: delayed but massive and lethal metastases of a chemotherapy-associated secondary malignancy. Urology 70 (1): 178.e3-6, 2007.
-
Thorner PS, Shago M, Marrano P, et al.: TFE3-positive renal cell carcinomas are not always Xp11 translocation carcinomas: Report of a case with a TPM3-ALK translocation. Pathol Res Pract 212 (10): 937-942, 2016.
-
Cajaiba MM, Jennings LJ, Rohan SM, et al.: ALK-rearranged renal cell carcinomas in children. Genes Chromosomes Cancer 55 (5): 442-51, 2016.
-
Smith NE, Deyrup AT, Mariño-Enriquez A, et al.: VCL-ALK renal cell carcinoma in children with sickle-cell trait: the eighth sickle-cell nephropathy? Am J Surg Pathol 38 (6): 858-63, 2014.
-
Cajaiba MM, Jennings LJ, George D, et al.: Expanding the spectrum of ALK-rearranged renal cell carcinomas in children: Identification of a novel HOOK1-ALK fusion transcript. Genes Chromosomes Cancer 55 (10): 814-7, 2016.
-
Imbalzano AN, Jones SN: Snf5 tumor suppressor couples chromatin remodeling, checkpoint control, and chromosomal stability. Cancer Cell 7 (4): 294-5, 2005.
-
Eaton KW, Tooke LS, Wainwright LM, et al.: Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer 56 (1): 7-15, 2011.
-
Versteege I, Sévenet N, Lange J, et al.: Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394 (6689): 203-6, 1998.
-
Schneppenheim R, Frühwald MC, Gesk S, et al.: Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet 86 (2): 279-84, 2010.
-
Hasselblatt M, Gesk S, Oyen F, et al.: Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am J Surg Pathol 35 (6): 933-5, 2011.
-
Lee RS, Stewart C, Carter SL, et al.: A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest 122 (8): 2983-8, 2012.
-
Biegel JA, Zhou JY, Rorke LB, et al.: Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res 59 (1): 74-9, 1999.
-
Biegel JA: Molecular genetics of atypical teratoid/rhabdoid tumor. Neurosurg Focus 20 (1): E11, 2006.
-
Bourdeaut F, Lequin D, Brugières L, et al.: Frequent hSNF5/INI1 germline mutations in patients with rhabdoid tumor. Clin Cancer Res 17 (1): 31-8, 2011.
-
Geller JI, Roth JJ, Biegel JA: Biology and Treatment of Rhabdoid Tumor. Crit Rev Oncog 20 (3-4): 199-216, 2015.
-
Janson K, Nedzi LA, David O, et al.: Predisposition to atypical teratoid/rhabdoid tumor due to an inherited INI1 mutation. Pediatr Blood Cancer 47 (3): 279-84, 2006.
-
Sévenet N, Sheridan E, Amram D, et al.: Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet 65 (5): 1342-8, 1999.
-
Hasselblatt M, Nagel I, Oyen F, et al.: SMARCA4-mutated atypical teratoid/rhabdoid tumors are associated with inherited germline alterations and poor prognosis. Acta Neuropathol 128 (3): 453-6, 2014.
-
Ueno-Yokohata H, Okita H, Nakasato K, et al.: Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat Genet 47 (8): 861-3, 2015.
-
Karlsson J, Valind A, Gisselsson D: BCOR internal tandem duplication and YWHAE-NUTM2B/E fusion are mutually exclusive events in clear cell sarcoma of the kidney. Genes Chromosomes Cancer 55 (2): 120-3, 2016.
-
Astolfi A, Melchionda F, Perotti D, et al.: Whole transcriptome sequencing identifies BCOR internal tandem duplication as a common feature of clear cell sarcoma of the kidney. Oncotarget 6 (38): 40934-9, 2015.
-
Roy A, Kumar V, Zorman B, et al.: Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat Commun 6: 8891, 2015.
-
O'Meara E, Stack D, Lee CH, et al.: Characterization of the chromosomal translocation t(10;17)(q22;p13) in clear cell sarcoma of kidney. J Pathol 227 (1): 72-80, 2012.
-
Gooskens SL, Gadd S, Guidry Auvil JM, et al.: TCF21 hypermethylation in genetically quiescent clear cell sarcoma of the kidney. Oncotarget 6 (18): 15828-41, 2015.
MelanomaMelanoma-related conditions with malignant potential that arise in the pediatric population can be classified into three general groups:[1] - Large/giant congenital melanocytic nevus.
- Spitzoid melanocytic tumors ranging from atypical Spitz tumors to spitzoid melanomas.
- Melanoma arising in older adolescents that shares characteristics with adult melanoma (i.e., conventional melanoma).
The genomic characteristics of each tumor are summarized in Table 3. The genomic landscape of conventional melanoma in children is represented by many of the genomic alterations found in adults with melanoma.[1] A report from the Pediatric Cancer Genome Project observed that 15 cases of conventional melanoma had a high burden of somatic single-nucleotide variations, TERT promoter mutations (12 of 13), and activating BRAF V600 mutations (13 of 15), as well as a mutational spectrum signature consistent with ultraviolet light damage. In addition, two-thirds of the cases had MC1R variants associated with an increased susceptibility to melanoma. The genomic landscape of spitzoid melanomas is characterized by kinase gene fusions involving various genes including RET, ROS1, NTRK1, ALK, MET, and BRAF.[2,3,4] These fusion genes have been reported in approximately 50% of cases and occur in a mutually exclusive manner.[1,3]TERT promoter mutations are uncommon in spitzoid melanocytic lesions and were observed in only 4 of 56 patients evaluated in one series. However, each of the four cases with TERT promoter mutations experienced hematogenous metastases and died of their disease. This finding supports the potential of TERT promoter mutations in predicting aggressive clinical behavior in children with spitzoid melanocytic neoplasms, but further study is needed to define the role of wild-type TERT promoter status in predicting clinical behavior in patients with primary site spitzoid tumors. Large congenital melanocytic nevi are reported to have activating NRAS Q61 mutations with no other recurring mutations noted.[5] Somatic mosaicism for NRAS Q61 mutations has also been reported in patients with multiple congenital melanocytic nevi and neuromelanosis.[6] Table 3. Characteristics of Melanocytic Lesions| Tumor | Affected Gene |
|---|
| Melanoma | BRAF,NRAS,KIT, NF1 | | Spitzoid melanoma | Kinase fusions (RET,ROS,MET,ALK,BRAF,NTRK1) | | Spitz nevus | HRAS;BRAFandNRAS(uncommon) | | Acquired nevus | BRAF | | Dysplastic nevus | BRAF,NRAS | | Blue nevus | GNAQ | | Ocular melanoma | GNAQ | | Congenital nevi | NRAS | (Refer to the PDQ summary on Unusual Cancers of Childhood Treatment for information about the treatment of childhood melanoma.) References:
-
Lu C, Zhang J, Nagahawatte P, et al.: The genomic landscape of childhood and adolescent melanoma. J Invest Dermatol 135 (3): 816-23, 2015.
-
Wiesner T, He J, Yelensky R, et al.: Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun 5: 3116, 2014.
-
Lee S, Barnhill RL, Dummer R, et al.: TERT Promoter Mutations Are Predictive of Aggressive Clinical Behavior in Patients with Spitzoid Melanocytic Neoplasms. Sci Rep 5: 11200, 2015.
-
Yeh I, Botton T, Talevich E, et al.: Activating MET kinase rearrangements in melanoma and Spitz tumours. Nat Commun 6: 7174, 2015.
-
Charbel C, Fontaine RH, Malouf GG, et al.: NRAS mutation is the sole recurrent somatic mutation in large congenital melanocytic nevi. J Invest Dermatol 134 (4): 1067-74, 2014.
-
Kinsler VA, Thomas AC, Ishida M, et al.: Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J Invest Dermatol 133 (9): 2229-36, 2013.
Thyroid CancerStudies have shown subtle differences between the genetic profiling of childhood differentiated thyroid carcinomas and that of adult tumors. In one study, a higher prevalence of RET/PTC rearrangements was reported in pediatric papillary carcinoma (45%-65% in children vs. 3%-34% in adults).[1]BRAF V600E mutations are seen in more than 50% of adults with papillary thyroid carcinoma;[2] although it likely occurs in a similar frequency in pediatric patients, studies have revealed a wide variation in frequency of this mutation.[1,2,3,4] In children, the correlation between the genomic alteration and stage or prognosis has not been well defined. While two studies failed to show a correlation,[3,4] one study that included 55 pediatric thyroid carcinoma cases demonstrated a significant correlation between the presence of a BRAF V600E mutation and an increased risk of recurrence.[5] Differentiated thyroid carcinoma has been associated with germline DICER1 mutations and it is considered part of the DICER1 syndrome.[6] Table 4. Comparison of Thyroid Carcinoma Characteristics in Children and Adolescents and Adultsa| Characteristic | Children and Adolescents (%) | Adults (%) |
|---|
| a Yamashita et al.,[7]Nikita et al.,[4]and Alzahrani et al.[5] | | Histologic subtype: | | | | Papillary | 67-98 | 85-90 | | Follicular | 4-23 | <10 | | Medullary | 2-8 | 3 | | Poorly differentiated | <0.1 | 2-7 | | | | Gene rearrangements: | | | | RET/PTC | 21-87 | 0-35 | | NTRK 1 | 5-11 | 5-13 | | AKAP9-BRAF | 11 | 1 | | PAX8-PPARG | Unknown | 0-50 | | | | Point mutations: | | | | BRAF | 0-63 | 0-43 | | RASfamily | 0-16 | 25-69 | | GNAS | 0 | 11 | | TP53 | 0-23 | 0-20 | | TERT | 0 | 16 | | | | Other: | | | | Multicentric | 30-50 | 40-56 | | Lymph node involvement | 30-90 | 5-55 | | Extrathyroid extension | 24-51 | 16-46 | | Vascular invasion | <31 | 14-37 | | Distant metastases | 10-20 | 5-10 | (Refer to the PDQ summary on Unusual Cancers of Childhood Treatment for information about the treatment of childhood thyroid cancer.) References:
-
Ballester LY, Sarabia SF, Sayeed H, et al.: Integrating Molecular Testing in the Diagnosis and Management of Children with Thyroid Lesions. Pediatr Dev Pathol 19 (2): 94-100, 2016 Mar-Apr.
-
Rivkees SA, Mazzaferri EL, Verburg FA, et al.: The treatment of differentiated thyroid cancer in children: emphasis on surgical approach and radioactive iodine therapy. Endocr Rev 32 (6): 798-826, 2011.
-
Henke LE, Perkins SM, Pfeifer JD, et al.: BRAF V600E mutational status in pediatric thyroid cancer. Pediatr Blood Cancer 61 (7): 1168-72, 2014.
-
Nikita ME, Jiang W, Cheng SM, et al.: Mutational Analysis in Pediatric Thyroid Cancer and Correlations with Age, Ethnicity, and Clinical Presentation. Thyroid 26 (2): 227-34, 2016.
-
Alzahrani AS, Qasem E, Murugan AK, et al.: Uncommon TERT Promoter Mutations in Pediatric Thyroid Cancer. Thyroid 26 (2): 235-41, 2016.
-
Slade I, Bacchelli C, Davies H, et al.: DICER1 syndrome: clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J Med Genet 48 (4): 273-8, 2011.
-
Yamashita S, Saenko V: Mechanisms of Disease: molecular genetics of childhood thyroid cancers. Nat Clin Pract Endocrinol Metab 3 (5): 422-9, 2007.
Multiple Endocrine Neoplasia SyndromesThe most salient clinical and genetic alterations of the multiple endocrine neoplasia (MEN) syndromes are shown in Table 5. Table 5. Multiple Endocrine Neoplasia (MEN) Syndromes with Associated Clinical and Genetic Alterations| Syndrome | Clinical Features/Tumors | Genetic Alterations |
|---|
| MEN type 1: Werner syndrome[1] | Parathyroid | 11q13 (MEN1 gene) | | Pancreatic islets: | Gastrinoma | 11q13 (MEN1 gene) | | Insulinoma | | Glucagonoma | | VIPoma | | Pituitary: | Prolactinoma | 11q13 (MEN1 gene) | | Somatotrophinoma | | Corticotropinoma | | Other associated tumors (less common): | Carcinoid: bronchial and thymic | 11q13 (MEN1 gene) | | Adrenocortical | | Lipoma | | Angiofibroma | | Collagenoma | | MEN type 2A: Sipple syndrome | Medullary thyroid carcinoma | 10q11.2 (RET gene) | | Pheochromocytoma | | Parathyroid gland | | MEN type 2B | Medullary thyroid carcinoma | 10q11.2 (RET gene) | | Pheochromocytoma | | Mucosal neuromas | | Intestinal ganglioneuromatosis | | Marfanoid habitus | - Multiple endocrine neoplasia type 1 (MEN1) syndrome (Werner syndrome): MEN1 syndrome is an autosomal dominant disorder characterized by the presence of tumors in the parathyroid, pancreatic islet cells, and anterior pituitary. Diagnosis of this syndrome should be considered when two endocrine tumors listed in Table 5 are present.
A study documented the initial symptoms of MEN1 syndrome occurring before age 21 years in 160 patients.[2] Of note, most patients had familial MEN1 syndrome and were followed up using an international screening protocol. - Primary hyperparathyroidism. Primary hyperparathyroidism, the most common symptom, was found in 75% of patients, usually only in those with biological abnormalities. Primary hyperparathyroidism diagnosed outside of a screening program is extremely rare, most often presents with nephrolithiasis, and should lead the clinician to suspect MEN1.[2,3]
- Pituitary adenomas. Pituitary adenomas were discovered in 34% of patients, occurred mainly in females older than 10 years, and were often symptomatic.[2]
- Pancreatic neuroendocrine tumors. Pancreatic neuroendocrine tumors were found in 23% of patients. Specific diagnoses included insulinoma, nonsecreting pancreatic tumor, and Zollinger-Ellison syndrome. The first case of insulinoma occurred before age 5 years.[2]
- Malignant tumors. Four patients had malignant tumors (two adrenal carcinomas, one gastrinoma, and one thymic carcinoma). The patient with thymic carcinoma died before age 21 years from rapidly progressive disease.
Germline mutations of the MEN1 gene located on chromosome 11q13 are found in 70% to 90% of patients; however, this gene has also been shown to be frequently inactivated in sporadic tumors.[4] Mutation testing is combined with clinical screening for patients and family members with proven at-risk MEN1 syndrome.[5] It is recommended that screening for patients with MEN1 syndrome begin by the age of 5 years and continue for life. The number of tests or biochemical screening is age specific and may include yearly serum calcium, parathyroid hormone, gastrin, glucagon, secretin, proinsulin, chromogranin A, prolactin, and IGF-1. Radiologic screening should include a magnetic resonance imaging of the brain and computed tomography of the abdomen every 1 to 3 years.[6] - Multiple endocrine neoplasia type 2A (MEN2A) and multiple endocrine neoplasia type 2B (MEN2B) syndromes:
A germline activating mutation in the RET oncogene (a receptor tyrosine kinase) on chromosome 10q11.2 is responsible for the uncontrolled growth of cells in medullary thyroid carcinoma associated with MEN2A and MEN2B syndromes.[7,8,9] - MEN2A: MEN2A is characterized by the presence of two or more endocrine tumors (refer to Table 5) in an individual or in close relatives.[10]RET mutations in these patients are usually confined to exons 10 and 11.
- MEN2B: MEN2B is characterized by medullary thyroid carcinomas, parathyroid hyperplasias, adenomas, pheochromocytomas, mucosal neuromas, and ganglioneuromas.[10,11,12] The medullary thyroid carcinomas that develop in these patients are extremely aggressive. More than 95% of mutations in these patients are confined to codon 918 in exon 16, causing receptor autophosphorylation and activation.[13] Patients also have medullated corneal nerve fibers, distinctive faces with enlarged lips, and an asthenic Marfanoid body habitus.
A pentagastrin stimulation test can be used to detect the presence of medullary thyroid carcinoma in these patients, although management of patients is driven primarily by the results of genetic analysis for RET mutations.[13,14]
Guidelines for genetic testing of suspected patients with MEN2 syndrome and the correlations between the type of mutation and the risk levels of aggressiveness of medullary thyroid cancer have been published.[14,15] - Familial Medullary Thyroid Carcinoma: Familial medullary thyroid carcinoma is diagnosed in families with medullary thyroid carcinoma in the absence of pheochromocytoma or parathyroid adenoma/hyperplasia. RET mutations in exons 10, 11, 13, and 14 account for most cases.
The most-recent literature suggests that this entity should not be identified as a form of hereditary medullary thyroid carcinoma that is separate from MEN2A and MEN2B. Familial medullary thyroid carcinoma should be recognized as a variant of MEN2A, to include families with only medullary thyroid cancer who meet the original criteria for familial disease. The original criteria includes families of at least two generations with at least two, but less than ten, patients with RET germline mutations; small families in which two or fewer members in a single generation have germline RET mutations; and single individuals with a RET germline mutation.[14,16]
Table 6. Clinical Features of Multiple Endocrine Neoplasia Type 2 (MEN2) Syndromes| MEN2 Subtype | Medullary Thyroid Carcinoma | Pheochromocytoma | Parathyroid Disease |
|---|
| MEN2A | 95% | 50% | 20% to 30% | | MEN2B | 100% | 50% | Uncommon | (Refer to the PDQ summary on Unusual Cancers of Childhood Treatment for information about the treatment of childhood MEN syndromes.) References:
-
Thakker RV: Multiple endocrine neoplasia--syndromes of the twentieth century. J Clin Endocrinol Metab 83 (8): 2617-20, 1998.
-
Goudet P, Dalac A, Le Bras M, et al.: MEN1 disease occurring before 21 years old: a 160-patient cohort study from the Groupe d'étude des Tumeurs Endocrines. J Clin Endocrinol Metab 100 (4): 1568-77, 2015.
-
Romero Arenas MA, Morris LF, Rich TA, et al.: Preoperative multiple endocrine neoplasia type 1 diagnosis improves the surgical outcomes of pediatric patients with primary hyperparathyroidism. J Pediatr Surg 49 (4): 546-50, 2014.
-
Farnebo F, Teh BT, Kytölä S, et al.: Alterations of the MEN1 gene in sporadic parathyroid tumors. J Clin Endocrinol Metab 83 (8): 2627-30, 1998.
-
Field M, Shanley S, Kirk J: Inherited cancer susceptibility syndromes in paediatric practice. J Paediatr Child Health 43 (4): 219-29, 2007.
-
Thakker RV: Multiple endocrine neoplasia type 1 (MEN1). Best Pract Res Clin Endocrinol Metab 24 (3): 355-70, 2010.
-
Sanso GE, Domene HM, Garcia R, et al.: Very early detection of RET proto-oncogene mutation is crucial for preventive thyroidectomy in multiple endocrine neoplasia type 2 children: presence of C-cell malignant disease in asymptomatic carriers. Cancer 94 (2): 323-30, 2002.
-
Alsanea O, Clark OH: Familial thyroid cancer. Curr Opin Oncol 13 (1): 44-51, 2001.
-
Fitze G: Management of patients with hereditary medullary thyroid carcinoma. Eur J Pediatr Surg 14 (6): 375-83, 2004.
-
Puñales MK, da Rocha AP, Meotti C, et al.: Clinical and oncological features of children and young adults with multiple endocrine neoplasia type 2A. Thyroid 18 (12): 1261-8, 2008.
-
Skinner MA, DeBenedetti MK, Moley JF, et al.: Medullary thyroid carcinoma in children with multiple endocrine neoplasia types 2A and 2B. J Pediatr Surg 31 (1): 177-81; discussion 181-2, 1996.
-
Brauckhoff M, Gimm O, Weiss CL, et al.: Multiple endocrine neoplasia 2B syndrome due to codon 918 mutation: clinical manifestation and course in early and late onset disease. World J Surg 28 (12): 1305-11, 2004.
-
Sakorafas GH, Friess H, Peros G: The genetic basis of hereditary medullary thyroid cancer: clinical implications for the surgeon, with a particular emphasis on the role of prophylactic thyroidectomy. Endocr Relat Cancer 15 (4): 871-84, 2008.
-
Waguespack SG, Rich TA, Perrier ND, et al.: Management of medullary thyroid carcinoma and MEN2 syndromes in childhood. Nat Rev Endocrinol 7 (10): 596-607, 2011.
-
Kloos RT, Eng C, Evans DB, et al.: Medullary thyroid cancer: management guidelines of the American Thyroid Association. Thyroid 19 (6): 565-612, 2009.
-
Wells SA Jr, Asa SL, Dralle H, et al.: Revised American Thyroid Association guidelines for the management of medullary thyroid carcinoma. Thyroid 25 (6): 567-610, 2015.
Changes to this Summary (04 / 25 / 2017)The PDQ cancer information summaries are reviewed regularly and updated as new information becomes available. This section describes the latest changes made to this summary as of the date above. Leukemias Revised text in the Acute Myeloid Leukemia (AML) section to state that both RUNX1-RUNX1T1 and CBFB-MYH11 subtypes commonly show mutations in genes that activate receptor tyrosine kinase signaling; NRAS and KIT are the most commonly mutated genes for both subtypes. Also added text to state that KIT mutations may indicate increased risk of treatment failure for patients with core-binding factor AML, although the prognostic significance of KIT mutations may be dependent on the mutant-allele ratio and/or the specific type of mutation. KIT exon 17 mutations are enriched in patients with RUNX1-RUNX1T1 fusions when compared with patients with CBFB-MYH11 fusions (cited Faber et al. and Eisfeld et al. as references 137 and 138, respectively). Revised text in the Acute Myeloid Leukemia (AML) section to state that cases with CBFB-MYH11 and cases with RUNX1-RUNX1T1 have distinctive secondary mutations; CBFB-MYH11 secondary mutations are primarily restricted to genes that activate receptor tyrosine kinase signaling. Central Nervous System Tumors The Diffuse Midline Glioma, H3 K27M-Mutant (Including Diffuse Intrinsic Pontine Gliomas [DIPGs]) section was extensively revised. Added text to the Central Nervous System (CNS) Atypical Teratoid/Rhabdoid Tumors (AT/RT) section to state that the 2016 World Health Organization (WHO) classification defines AT/RT by the presence of either SMARCB1 or SMARCA4 alterations. Tumors with histological features of AT/RT that lack these genomic alterations are termed CNS embryonal tumor with rhabdoid features (cited Louis et al. as reference 1). Added text to the Central Nervous System (CNS) Atypical Teratoid/Rhabdoid Tumors (AT/RT) section to state that for patients with AT/RT TYR, the mean overall survival (OS) is 37 months. Cribriform neuroepithelial tumor is a brain cancer that also presents in young children and has genomic and epigenomic characteristics that are very similar to AT/RT TYR (cited Johann et al. as reference 59). Added text to the Central Nervous System (CNS) Atypical Teratoid/Rhabdoid Tumors (AT/RT) section to state that for patients with AT/RT SHH, the mean OS is 16 months. Added text to the Central Nervous System (CNS) Atypical Teratoid/Rhabdoid Tumors (AT/RT) section to state that for patients with AT/RT MYC, the mean OS is 13 months. Added text to the Ependymomas section to state that the 2016 WHO classification has accepted ependymoma, RELA fusion-positive, as a distinct diagnostic entity. Sarcomas Added text to the Ewing Sarcoma section to state that a detailed analysis of 85 patients with small round blue cell tumors that were negative for EWSR1 rearrangement by fluorescence in situ hybridization (FISH) with an EWSR1 break-apart probe identified eight patients with FUS rearrangements. Four patients who had EWSR1-ERG fusions were not detected by FISH with an EWSR1 break-apart probe. The authors do not recommend relying solely on EWSR1 break-apart probes for analyzing small round blue cell tumors with strong immunohistochemical positivity for CD99 (cited Chen et al. as reference 26). Langerhans Cell Histiocytosis Added text to the Langerhans Cell Histiocytosis section to state that in-frame BRAF deletions and in-frame FAM73A-BRAF fusions have occurred in the group of BRAF V600E and MAP2K1 mutation-negative cases (cited Chakraborty et al. as reference 12). Neuroblastoma Added text to the Neuroblastoma section about subclonal ALK mutations in neuroblastoma (cited Bellini et al. as reference 21). This summary is written and maintained by the PDQ Pediatric Treatment Editorial Board, which is editorially independent of NCI. The summary reflects an independent review of the literature and does not represent a policy statement of NCI or NIH. More information about summary policies and the role of the PDQ Editorial Boards in maintaining the PDQ summaries can be found on the About This PDQ Summary and PDQ® - NCI's Comprehensive Cancer Database pages. About This PDQ SummaryPurpose of This Summary This PDQ cancer information summary for health professionals provides comprehensive, peer-reviewed, evidence-based information about the genomics of childhood cancer. It is intended as a resource to inform and assist clinicians who care for cancer patients. It does not provide formal guidelines or recommendations for making health care decisions. Reviewers and Updates This summary is reviewed regularly and updated as necessary by the PDQ Pediatric Treatment Editorial Board, which is editorially independent of the National Cancer Institute (NCI). The summary reflects an independent review of the literature and does not represent a policy statement of NCI or the National Institutes of Health (NIH). Board members review recently published articles each month to determine whether an article should: - be discussed at a meeting,
- be cited with text, or
- replace or update an existing article that is already cited.
Changes to the summaries are made through a consensus process in which Board members evaluate the strength of the evidence in the published articles and determine how the article should be included in the summary. The lead reviewer for Childhood Cancer Genomics is: - Malcolm A. Smith, MD, PhD (National Cancer Institute)
Any comments or questions about the summary content should be submitted to Cancer.gov through the NCI website's Email Us. Do not contact the individual Board Members with questions or comments about the summaries. Board members will not respond to individual inquiries. Levels of Evidence Some of the reference citations in this summary are accompanied by a level-of-evidence designation. These designations are intended to help readers assess the strength of the evidence supporting the use of specific interventions or approaches. The PDQ Pediatric Treatment Editorial Board uses a formal evidence ranking system in developing its level-of-evidence designations. Permission to Use This Summary PDQ is a registered trademark. Although the content of PDQ documents can be used freely as text, it cannot be identified as an NCI PDQ cancer information summary unless it is presented in its entirety and is regularly updated. However, an author would be permitted to write a sentence such as "NCI's PDQ cancer information summary about breast cancer prevention states the risks succinctly: [include excerpt from the summary]." The preferred citation for this PDQ summary is: PDQ® Pediatric Treatment Editorial Board. PDQ Childhood Cancer Genomics. Bethesda, MD: National Cancer Institute. Updated <MM/DD/YYYY>. Available at: https://www.cancer.gov/types/childhood-cancers/pediatric-genomics-hp-pdq. Accessed <MM/DD/YYYY>. [PMID: 27466641] Images in this summary are used with permission of the author(s), artist, and/or publisher for use within the PDQ summaries only. Permission to use images outside the context of PDQ information must be obtained from the owner(s) and cannot be granted by the National Cancer Institute. Information about using the illustrations in this summary, along with many other cancer-related images, is available in Visuals Online, a collection of over 2,000 scientific images. Disclaimer Based on the strength of the available evidence, treatment options may be described as either "standard" or "under clinical evaluation." These classifications should not be used as a basis for insurance reimbursement determinations. More information on insurance coverage is available on Cancer.gov on the Managing Cancer Care page. Contact Us More information about contacting us or receiving help with the Cancer.gov website can be found on our Contact Us for Help page. Questions can also be submitted to Cancer.gov through the website's Email Us. Last Revised: 2017-04-25 Last modified on: 8 September 2017
|
|